I have asked the question on MSE as well: https://mathematica.stackexchange.com/questions/146550
I have the following image that i segment into respective cells using the function below:
segmentImage[binarizedMask_?ImageQ] :=
Module[{seg, areas, indexMaxarea, maxArea},
seg = MorphologicalComponents@*ColorNegate@Dilation[binarizedMask, 1];
areas = ComponentMeasurements[seg, "Area"];
{indexMaxarea, maxArea} = First@MaximalBy[areas, Last] /. Rule -> List;
If[maxArea > 20000, ArrayComponents[seg, Length@areas, indexMaxarea -> 0], seg]];
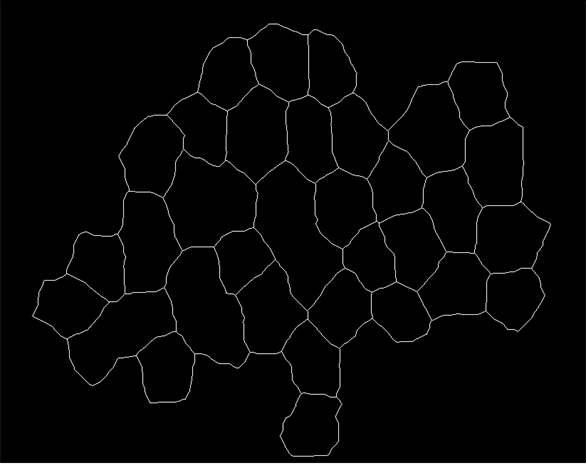
The result of the segmentation img2 = segmentImage[img]//Colorize is shown below:
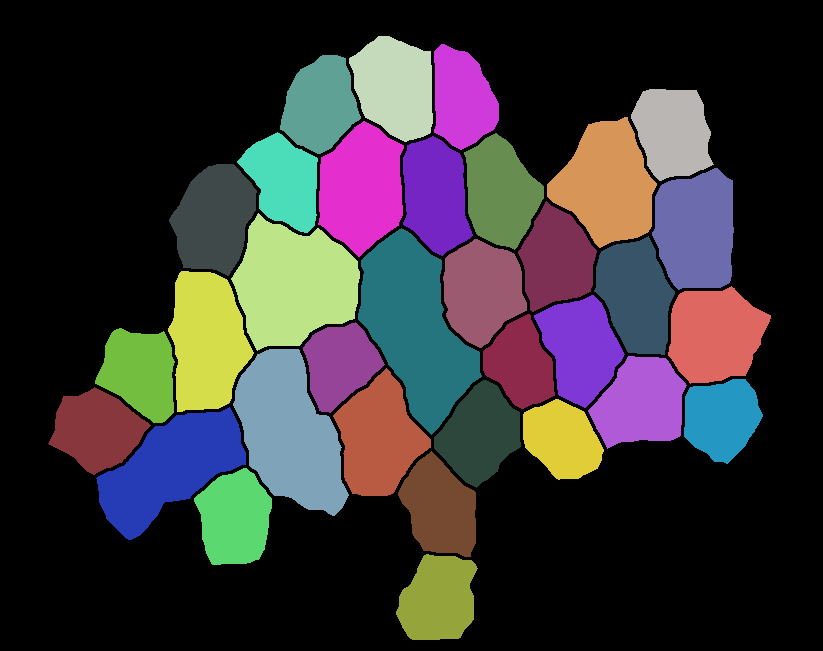
The problem that I am facing is that the boundaries between the cells are completely lost. I cannot recover any information of the neighbours or the neighbourcount:
ComponentMeasurements[img2,"NeighborCount"]
(* {1 -> 0, 2 -> 0, 3 -> 0, 4 -> 0, 5 -> 0, 6 -> 0, 7 -> 0, 8 -> 0,
9 -> 0, 10 -> 0, 11 -> 0, 12 -> 0, 13 -> 0, 14 -> 0, 15 -> 0,
16 -> 0, 17 -> 0, 18 -> 0, 19 -> 0, 20 -> 0, 21 -> 0, 22 -> 0,
23 -> 0, 24 -> 0, 25 -> 0, 26 -> 0, 27 -> 0, 28 -> 0, 29 -> 0,
30 -> 0, 31 -> 0, 32 -> 0, 33 -> 0} *)
This should not be the case since in the original image the cells are sharing boundaries. Is there a way to recover the neighbours or better way to do segmentation so that I do not lose boundaries between the cells?