I have three images image1, image2 and image3 from a time-lapse video obtained from an optical microscope. The cells move a little from their positions and new cells can enter and old cells can leave the Field of View. The binarized masks of the cell contours are posted below
The images are posted below: image1 
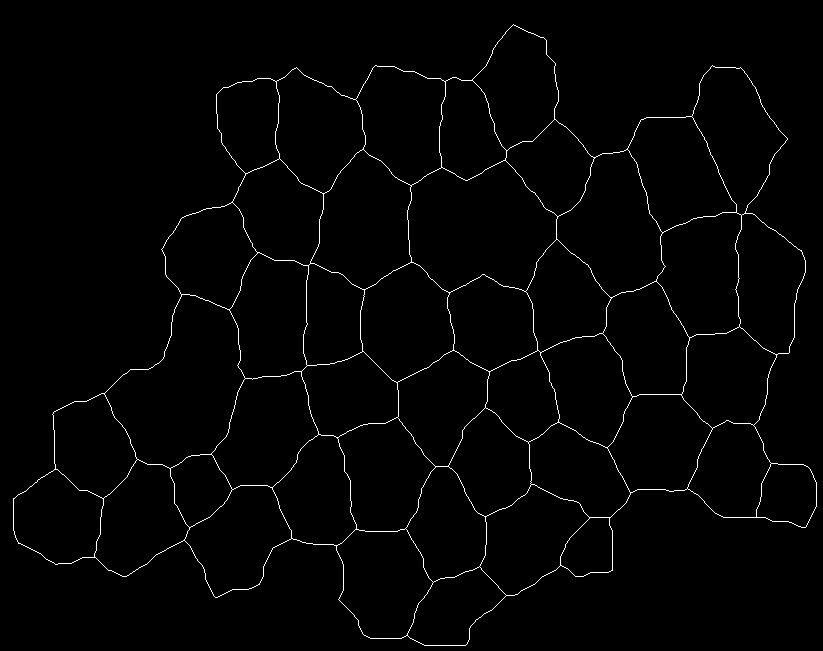
image2
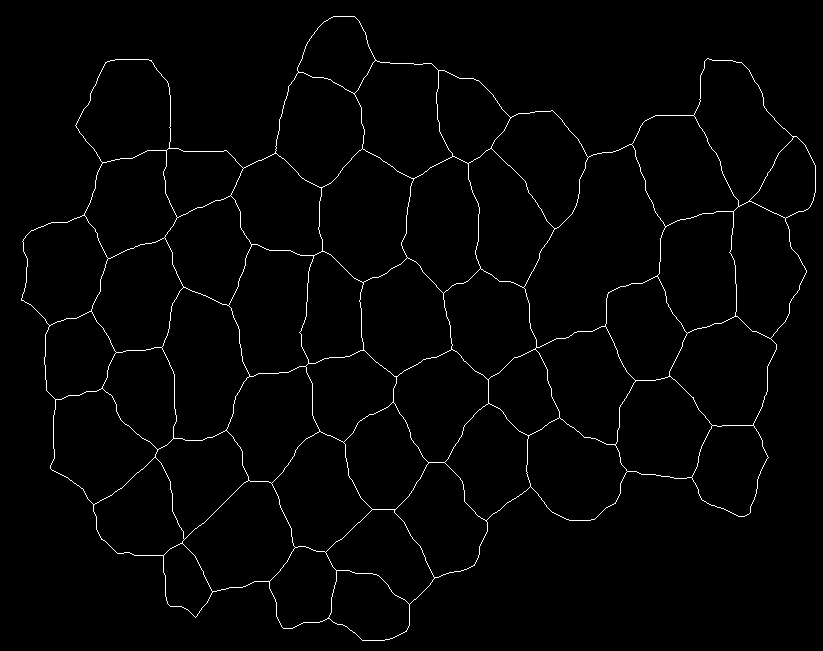
image3 is below:
We can segment the images using the function
segmentImage[binarizedMask_?ImageQ, threshCellsize_: 20000] :=
Module[{seg, areas, indexMaxarea, maxArea},
seg = MorphologicalComponents@*ColorNegate@
Dilation[binarizedMask, 1];
areas = ComponentMeasurements[seg, "Area"];
{indexMaxarea, maxArea} =
First@MaximalBy[areas, Last] /. Rule -> List;
If[maxArea > threshCellsize,
ArrayComponents[seg, Length@areas, indexMaxarea -> 0], seg] ~
Dilation~1];
If you closely look at the first image and the second image one can see that some cells have been incorrectly merged together when they should have been separate.
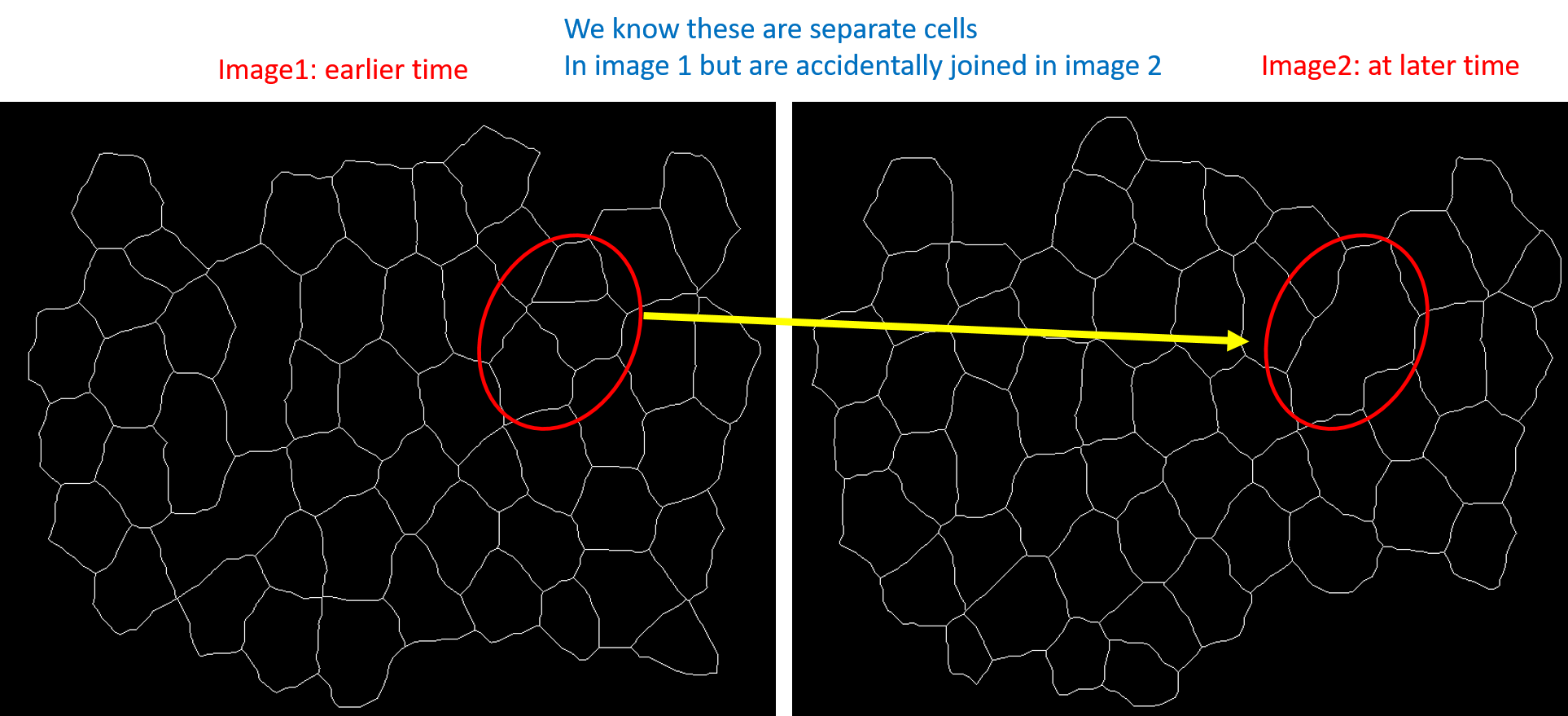
I wanted to construct a scheme to use image1 to correct for any incorrect merging in image2 and the corrected image (the result of correcting image2) to be used for correcting any merging in image3.
For this purpose i first compute an overlapMatrix (this matrix tells us how much area overlap exists between the different cells in two images). I am using the function proposed here by @kglr:
https://mathematica.stackexchange.com/questions/146558
Clear@overlapMatrix;
overlapMatrix[segPrev_, segCurr_] := Module[{labelPrev, maxLabelCurr},
labelPrev = Values@ComponentMeasurements[segPrev, "Label"];
maxLabelCurr =
MaximalBy[ComponentMeasurements[segCurr, "Label"], Value] /.
HoldPattern[_ -> x_] :> x;
Normal[SparseArray[SparseArray[#, maxLabelCurr] &@
DeleteCases[
Normal@*Counts@Flatten[(1 - Unitize[segPrev - #]) segCurr],
0 -> _] & /@ labelPrev]]
];
using the segmented images and the overlap matrix i can identify where merging is taking place using identifyFusions
Clear@identifyFusions;
identifyFusions[currSeg_, prevSeg_, overlapMat_,
overlapThresh_: 1200] :=
With[{dim = First@*Dimensions@prevSeg},
Module[{pos, candidates, isFusion},
isFusion[currInd_, prevInd_List] :=
Module[{mask, candCentroid, trueList},
mask = ComponentMeasurements[currSeg, "Mask"][[currInd, 2]];
candCentroid =
ComponentMeasurements[prevSeg, "Centroid"][[prevInd, 2]];
candCentroid =
Map[{dim - #[[1]], #[[2]]} &@*Round@*Reverse, candCentroid];
And @@ (MemberQ[ArrayRules@mask, # -> 1] & /@ candCentroid)
];
pos = Position[overlapMat, x_ /; x > overlapThresh];
candidates = Cases[Normal@GroupBy[pos, Last -> First],
HoldPattern[_ -> {Repeated[_, {2, \[Infinity]}]}]];
Pick[candidates, KeyValueMap[isFusion, <|candidates|>], True]
]
];
once i have identified incorrect fusions/merging of cells i can use a function breakFusion to separate the incorrectly fused cells
Clear@breakingFusion;
breakingFusion[currSeg_, prevSeg_][rule_] :=
Module[{maxLabelCurr, maxlabel, blobind, cluster, tempSeg,
unassignedpos, t, i = 1, nf, replaceExtraneous, blobpos, indpos, masks},
maxLabelCurr = maxlabel = Max@Values@ComponentMeasurements[currSeg, "Label"];
{blobind, cluster} = rule /. Rule -> List;
blobpos = Position[currSeg, blobind];
tempSeg = Fold[ReplacePart[#, (t[i++] = (blobpos \[Intersection]
Position[prevSeg, #2])) -> ++maxLabelCurr] &, currSeg, cluster];
unassignedpos = Position[tempSeg, blobind];
nf = Nearest@Flatten[MapThread[Thread[#1 -> #2] &, {Array[t, i - 1],
Range[maxlabel + 1, maxLabelCurr]}]];
replaceExtraneous = Replace[Map[# -> nf[#, 1] &, unassignedpos],HoldPattern[
p : {_, _} -> {x_}] :> p -> x, {1}];
ReplacePart[tempSeg, replaceExtraneous]
];
Now combining everything below in stackCorrection
stackCorrection[segmentPrev_, imageCurr_] :=
Module[{overlaps, segmentCurr, fusions},
segmentCurr = segmentImage[imageCurr]; (* segmentation *)
overlaps =
overlapMatrix[segmentPrev, segmentCurr]; (*compute overlaps*)
fusions = identifyFusions[segmentCurr, segmentPrev, overlaps]; (*
identify fusions *)
If[fusions != {},
segmentCurr = ArrayComponents@Fold[breakingFusion[#, segmentPrev][#2] &, segmentCurr,
fusions]] (* replace the fusions by breaking the cluster *)
];
The scheme mentioned above can be called:
correction = FoldList[stackCorrection, segmentImage@image1, {image2}];
correction[[2]] (* the corrected version of `image2` using `image1` *)
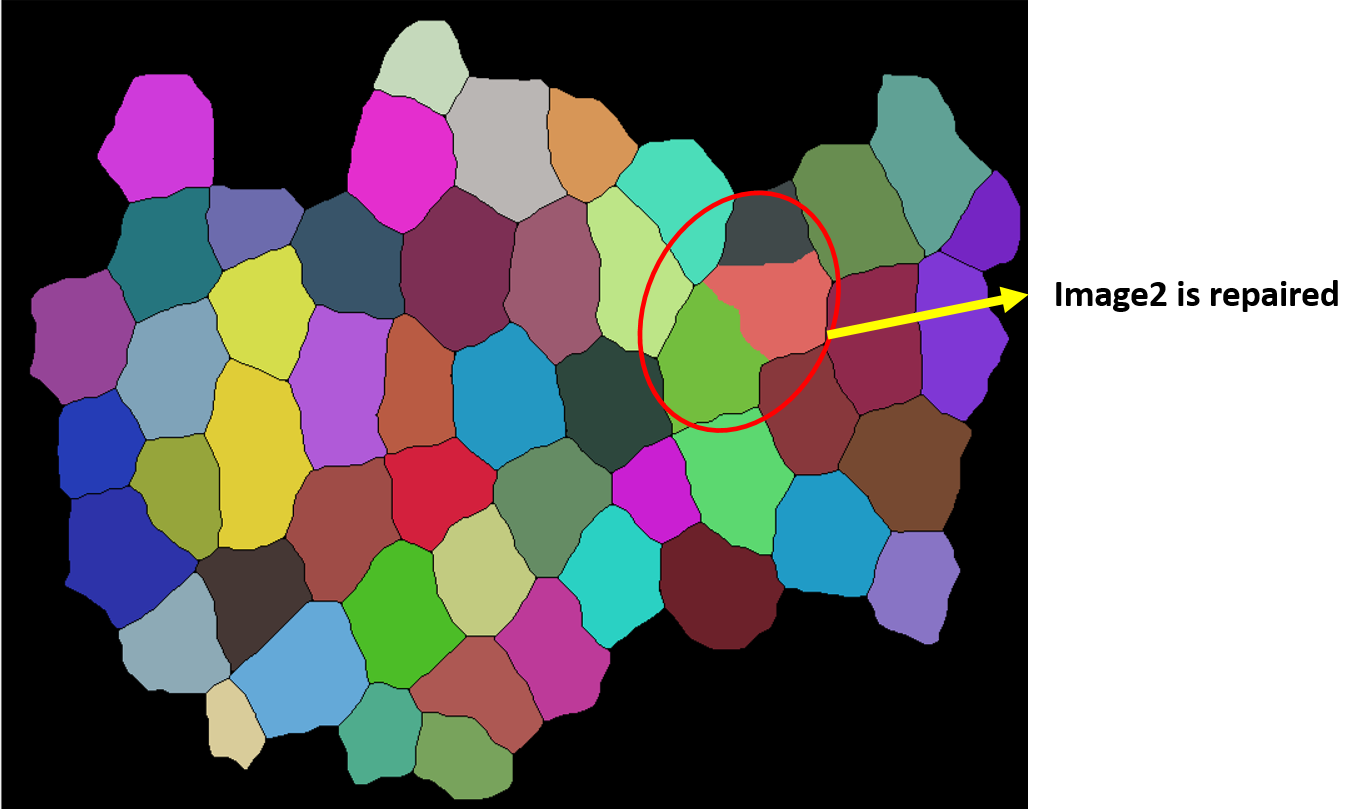
As we can see this works fine !
The Problem
when i instead use the corrected mask (obtained from image1 and image2) for further correcting image3 in the FoldList implementation:
correction = FoldList[stackCorrection, segmentImage@image1, {image2, image3}]
i get errors.
I should not be getting errors because two of the three cells (circled in image1) are incorrectly merged in image3.
However, the combinations below work e.g.
correction = FoldList[stackCorrection, segmentImage@image1, {image3}]
correction = FoldList[stackCorrection, segmentImage@image2, {image3}]
It is only when i give multiple images as the second argument that i encounter problem. Any help will be very much appreciated.