During the Wolfram Summer School Armenia 2016, I worked on the project Facial Emotion Recognition, to determine the emotions on faces dynamically. The successful outcome of the project made me think about whether it is possible to do the same procedure on a medical problem. After studying different problems, I ended up with breast cancer diagnosis in mammograms. Before getting into the technical side of the problem, it's better to know more of breast cancer:
Breast cancer is the most common cancer among american women, after skin cancers. About 1 in 8 (12%) of U.S. women will develop invasive breast cancer during their lifetime. But advances in breast cancer treatment mean many women can expect to beat the disease and maintain their physical appearance. 5-Year survival rate for women with breast cancer was 89% in 2015, up from the 63% in 1960s. (Read more)
Breast cancer is sometimes found after symptoms appear, but many women with breast cancer have no symptoms. Different tests can be used to look for and diagnose breast cancer. If your doctor finds an area of concern on a screening test (a mammogram), or if you have symptoms that could mean breast cancer, you will need more tests to know for sure if its cancer. After all, a biopsy is the only way to know FOR SURE if its cancer. (Read more) However, knowing that we are facing a complex problem, we can continue to the technical part of it:
Thanks to University of Southern California, we are using DDSM (Digital Database for Screening Mammography) for this projects. Data is divided to 3 groups of Normal, Benign and Cancer which represents the case condition. The Normal group contained 1624 samples, Benign and Cancer did 181 and 283 respectively. (Unfortunately, not great amount of data in Machine Learning scales.) At last all images needed to be converted to PNG format before starting to code.
Tolerance problem
Reviewing the images, we noticed that there is no strict discipline in positioning in the mammography. many of them were unadjusted and some reflected right to left. To gain a more reliable system, all images were rotated by 5, 10 and 15 in both clockwise and counterclockwise directions (appropriate cut-offs were performed on newly generated artifacts) and even been reflected from left to right. The manual generation increased overall number of the images by 14 times.
rotateDatas[x_] := List[
x
,
ImageRotate[x, -5 Degree, Background -> Black]
,
ImageRotate[x, +5 Degree, Background -> Black]
,
ImageRotate[x, -10 Degree, Background -> Black]
,
ImageRotate[x, +10 Degree, Background -> Black]
,
ImageRotate[x, -15 Degree, Background -> Black]
,
ImageRotate[x, +15 Degree, Background -> Black]
];
reflectData := List[Map[(ImageReflect[#, Right -> Left]) &, #], #] &;
Neural network
Like the Facial Emotion Recognition project, LeNet5 network was used for this project. The encoder layer has 3 classes: Normal, Benign, Cancer.
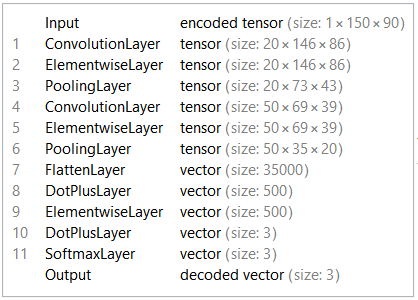
Training
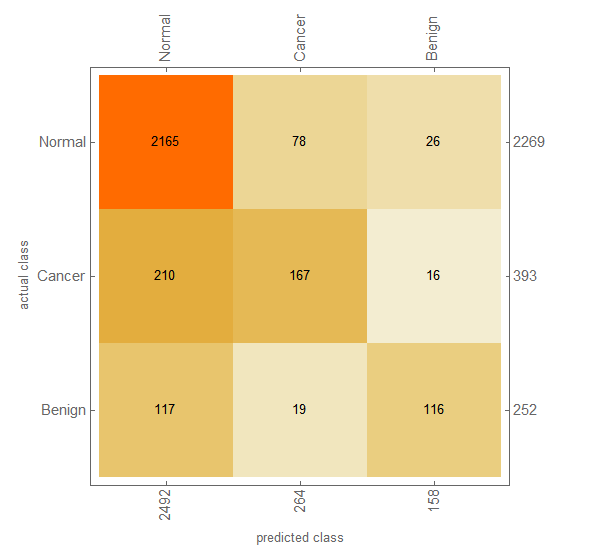
80% of the data is randomly selected as training set. Half of the remaining images (10%) is used as test data and the rest (10%) as validation data. It would have taken 24 hours to train the network on "Intel Core i5 4210U CPU, Windows 10 x64" for 20 rounds. But using my GPU (NVIDIA GeForce 840M), this time reduced to about an hour. (Thanks to Wolfram team!) However, training stopped sooner due to over-fitting. Applying the test set to the network, resulted this confusion matrix:
Issues and Further Improvement
The systems does well in Normal data, But not in Cancer and Benign cases. I see two major problems in it:
Assume an extreme case, the network estimates 51% of being Normal and 49% of Cancer. The encoding layer assumes it as Normal. A radiologist usually asks for further tests if there is the slightest chance of being non-normal.
Assume another extreme case, that the networks estimates 34% of being Normal and 33% for each Benign and Cancer. Again it will be perceived as Normal.
It's better to have a network to estimate the chance of being non-normal and warn the patient if the chances were higher than a threshold (20%, 30% or whatever). If yes, the case should be given to a specific network which decides whether it is Cancer or Benign.
After all, more data is always a mercy.
You can find data sets, network file and the notebook, here. You have to put the MX files in $HomeDirectory (usually Documents folder for Windows users) to evaluate it.