GOAL OF THE PROJECT: Understand the behavior of entanglement codes using simple computational models like cellular automata.
Erasure codes are a common approach for storing data redundantly to provide secure and long-term data storage. The motivation for this study is to understand the failure repair dynamics of entanglement codes.
Entanglement codes are designed for high fault-tolerance. They can provide high levels of availability and durability even in catastrophic scenarios. The redundancy is created and propagated through the system by tangling new data blocks with old ones, building entangled data chains that are woven into a growing mesh of interdependent content.
This project is about the development of a research framework to study failure repair dynamics on storage systems. The program allows playing with code settings and distinct failure assumptions. This post explains how simple entanglement codes work. In the next posts, I will explain complex entanglements and some of the results achieved during WSS17.
Project preliminaries
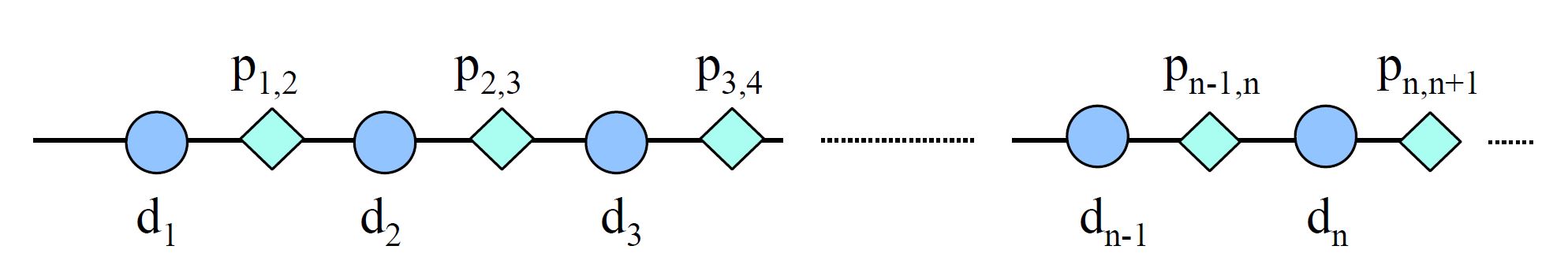
The above figure illustrates a simple entanglement code constructed with an open chain that alternates data blocks and parity blocks. , e.g, d2 is entangled with p1,2 and generates p2,3. Each new data block that is added to the system is XORed to the previous element.
Toy example: Hello, World
In this example, the word "Hello" is encoded with simple entanglements using Wolfram Language. The first step is to create a graph that shows the connections between encoded elements. To simplify the model, edges represent parities and vertices represent data blocks. Simple entanglement codes generate one parity for each data block added to the system. Therefore, we need at least 5 vertices to store the word hello and five edges to store the parities. Additionally, we need one vertex and one edge at the beginning of the chain. These additional elements only contain "zeros" but are needed to bootstrap the system. Finally, the chain ends with an additional vertex to connect the last generated parity.
g = Graph[{0 -> 1, 1 -> 2, 2 -> 3, 3 -> 4, 4 -> 5, 5 -> 6},
VertexLabels -> "Name", VertexSize -> Medium,
VertexStyle -> {0 -> Red, 6 -> Red}]
Write data: Now, we write "Hello" by inserting the character codes in the vertices:
vertexValues = ToCharacterCode["Hello"]
vertexValues = Insert[vertexValues, 0, {{1}, {-1}}]
vertexValues = {0, 72, 101, 108, 108, 111, 0}
Encode data: We can generate each parity individually by XORing the last two elements of the chain. But we can also encode all data simultaneously with the help of FoldList function:
encodedData = FoldList[BitXor, 0, vertexValues]
encodedData = {0, 72, 45, 65, 45, 66}
Decode data: We can read the content of a node by applying the bitwise XOR to the weight of its two adjacent edges. For example, to read "e" (char code 101) we use the adjacent edges that contain 72 and 45.
FromCharacterCode[BitXor[encodedData[[2]], encodedData[[3]]]]
e
Decode parities: There are two ways to recompute the content of a parity according to the direction we are moving on the chain. In this example, we recomputed encodedData[[3]]
Moving from left to right:
FromCharacterCode[BitXor[encodedData[[2]], vertexValues[[3]]]]
Moving from right to left:
FromCharacterCode[BitXor[encodedData[[4]], vertexValues[[4]]]
To visualize the graph:
g = Graph[{0 -> 1, 1 -> 2, 2 -> 3, 3 -> 4, 4 -> 5, 5 -> 6},
VertexSize -> Medium,
VertexStyle -> {0 -> Red, 6 -> Red},
VertexWeight -> Map[FromCharacterCode, vertexValues],
EdgeWeight -> Map[FromCharacterCode, encodedData],
VertexLabels -> "VertexWeight",
EdgeLabels -> "EdgeWeight"]

In the next post, I will discuss complex entanglements. Stay tuned!
 Attachments:
Attachments: