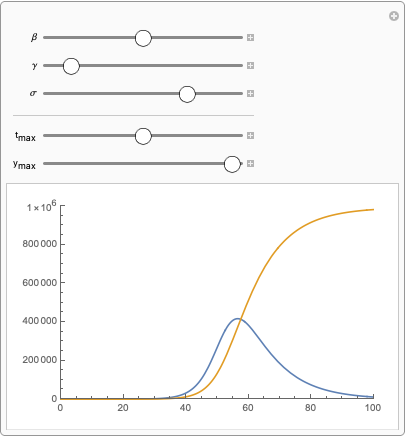
Hi Ravi,
It is easier to pick the solutions you want to plot from the full list. They are in S, E, I, R order, I and R are the third and fourth.
With[{seirSolutions =
seirCompartments /.
ParametricNDSolve[seirEquations /. seirInitialConditions, seirCompartments, {t, 0, 100000},
{\[Beta], \[Gamma], \[Sigma]}]},
Manipulate[
Show[{Plot[
Evaluate@
Through[seirSolutions[\[Beta], \[Gamma], \[Sigma]]][[3 ;; 4]], {t, 0, tmax},
PlotRange -> {{0, tmax}, {0, ymax}}]}],
(*paramters in model*)
{{\[Beta], 1/2}, 0, 1}, {{\[Gamma], 1/10}, 0, 1}, {{\[Sigma], 3/4}, 0, 1},
Delimiter,
(*plot range limits*)
{{tmax, 100, "\!\(\*SubscriptBox[\(t\), \(max\)]\)"}, 0, 200},
{{ymax, 1000000, "\!\(\*SubscriptBox[\(y\), \(max\)]\)"}, 0, 1000000},
SaveDefinitions -> True]]