Edit: I have added more functionality to the code: now the code is also able to track cells that merge to form colonies. The .m files below have been updated with new files. The github repository has the modified files as well.
Biological cells are active entities that undergo complex morphological transformation, migration and/or division to generate new daughter cells. For biologists and biophysicists it is important to understand the dynamics of individual cells as well as the cell-cell interactions that pave way for driving self-assembly/organization starting from the microscopic, to the mesoscopic and ultimately, the macroscopic scale - patterning the whole organism.
In order to map such intricate interactions quantitatively, we often need to track biological cells across time-lapse images obtained from either video or fluorescent microscopy. Such tracking is done using a series of binarized image masks. The dynamic characteristics of cells - within a tissue - makes tracking them a non-trivial process. The ability of cells to swap their immediate neighbours in a randomized manner can further complexify the process. Furthermore, in a high density setting, neighbouring cells can come so close together that the binarized mask might represent them incorrectly as a single entity (a fused object).
While there are a plethora of algorithms available to perform cell-tracking, most are not robust enough. Let me explain. A simple scheme for cell-tracking requires a segmented mask (a matrix where all the pixels of a specific cell has a unique label) as input from the user. The segmented mask itself can be generated by utilizing various image-processing algorithms and steps. As is often the case, there is a feedback between the tracking and the segmentation scheme. Nevertheless, the two schemes should be decoupled to ensure robustness of the tracking scheme.
The Wolfram Language implementation of the overlap-based cell tracker is based on an article by Chalfoun et al, Scientific Reports, 2016 (https://www.nature.com/articles/srep36984). The tracking algorithm can robustly track cells from a sequence of 2D images without any manual intervention. Furthermore, it corrects for any incorrect fusions/mergers of cells in an automated fashion and detects possible division (mitotic) events using stringent criterions for mother/daughter cells. The algorithm used for tracking is not specific to the segmentation scheme, which indeed makes lineage mapper more robust than many tracking schemes out there ! (Read the article for more detail). In short, the tracking scheme utilizes a minimization scheme over a cost matrix (the Hungarian Algorithm). In this implementation, I have used a graph theoretic approach to generate the same results.
The code below is the main tracking scheme (GitHub Code). For auxiliary files please refer to Github. Here are some of the results for tracking:
Input binarized mask from user -------------------- output segmentation and tracking
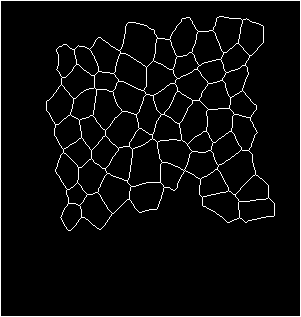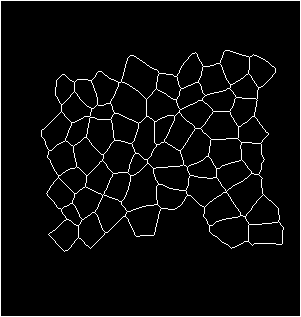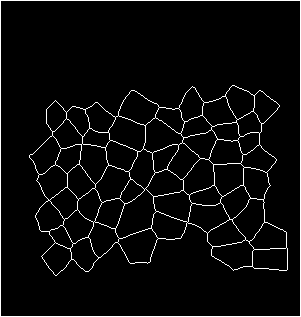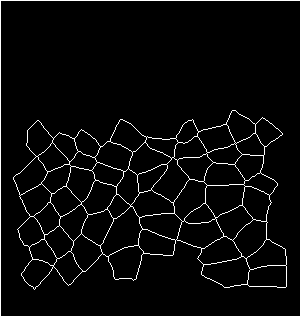
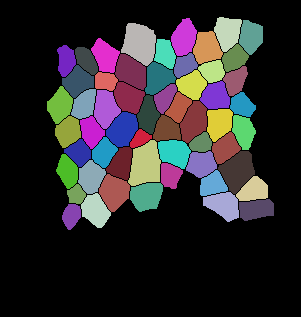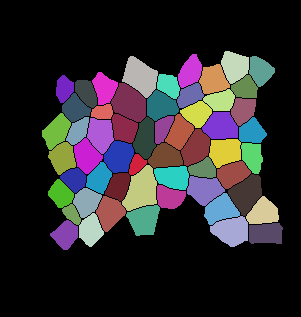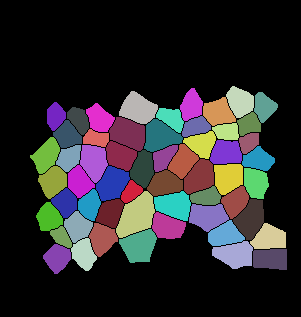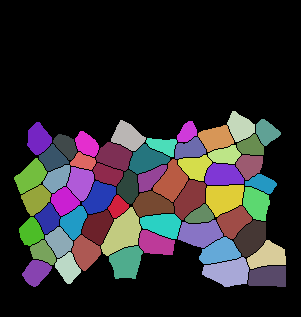
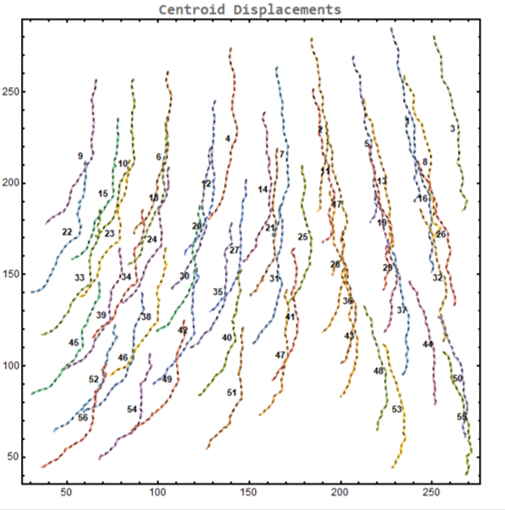
centroid displacement of tracked cells
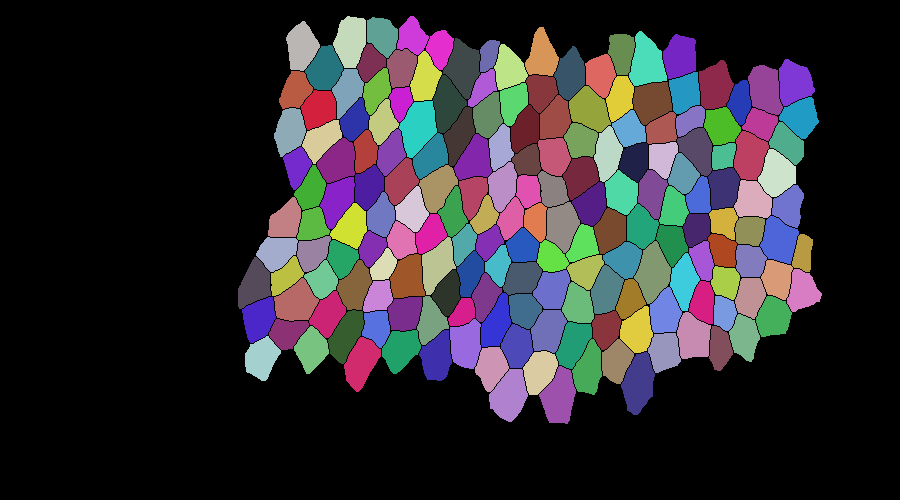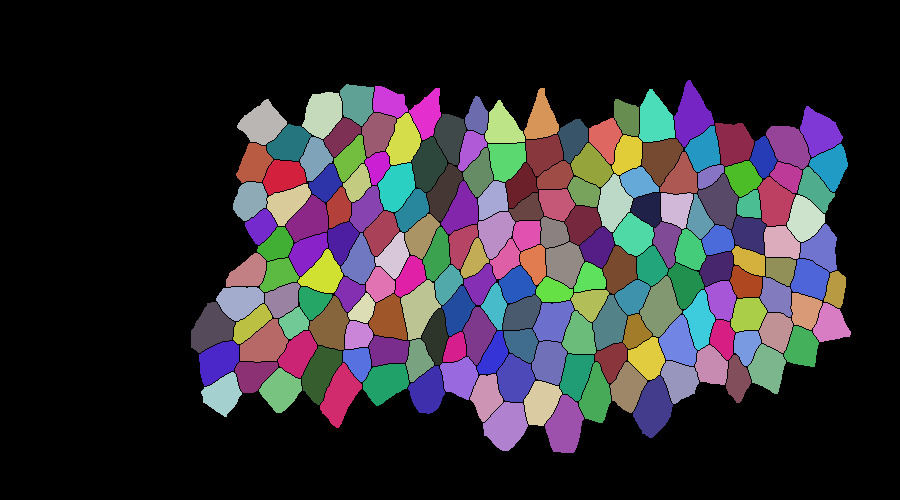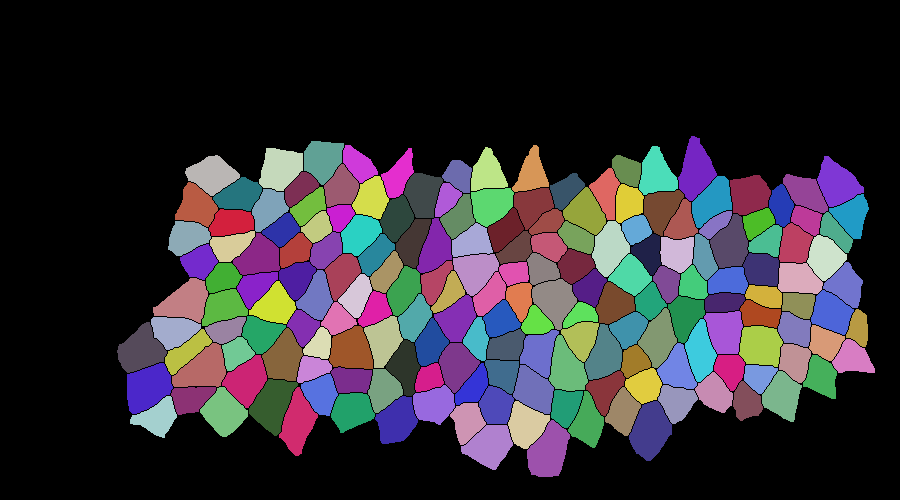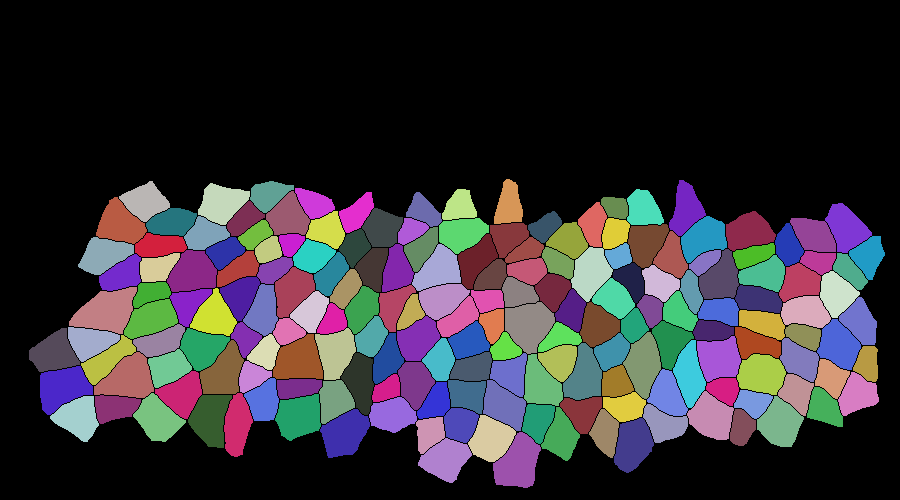
tracking a slightly bigger mask: evaluation took 32 seconds for 40 frames with 172 cells
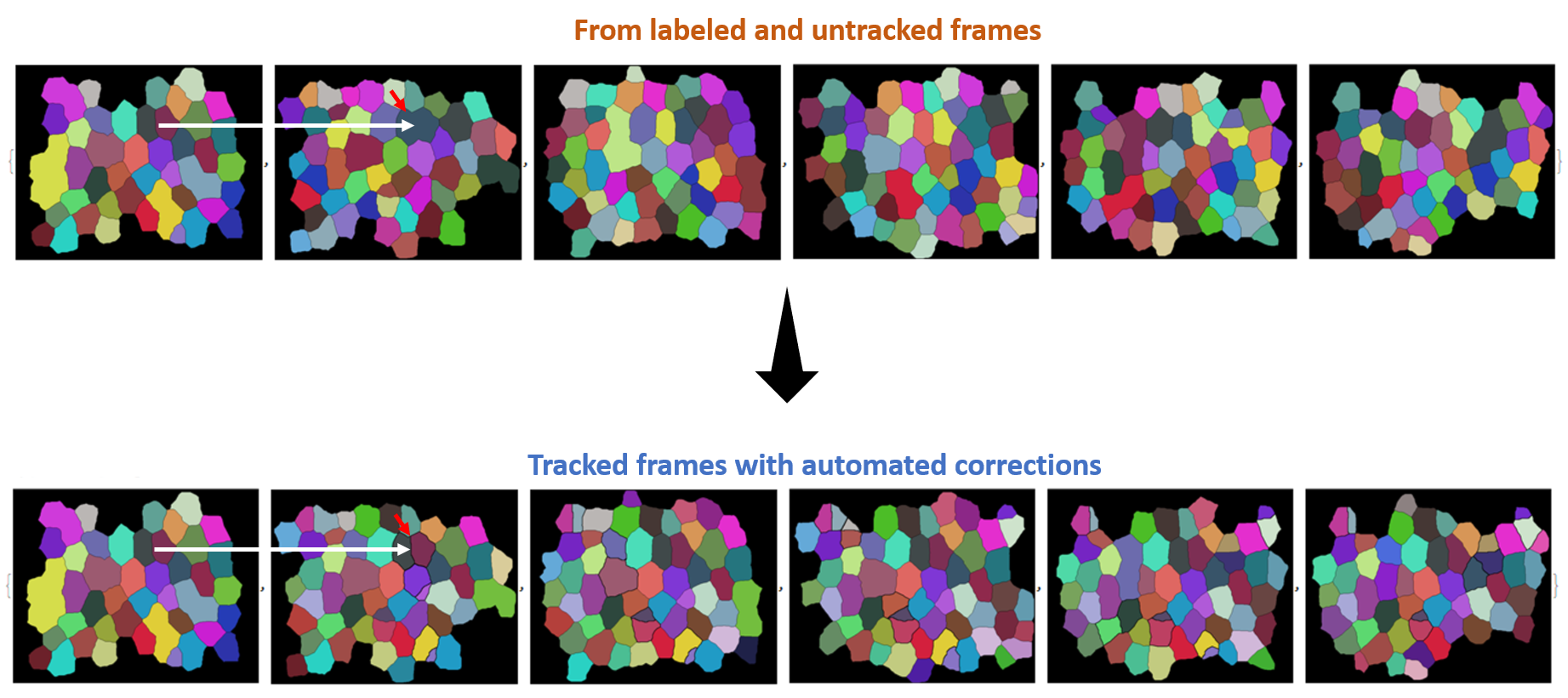
mask correction
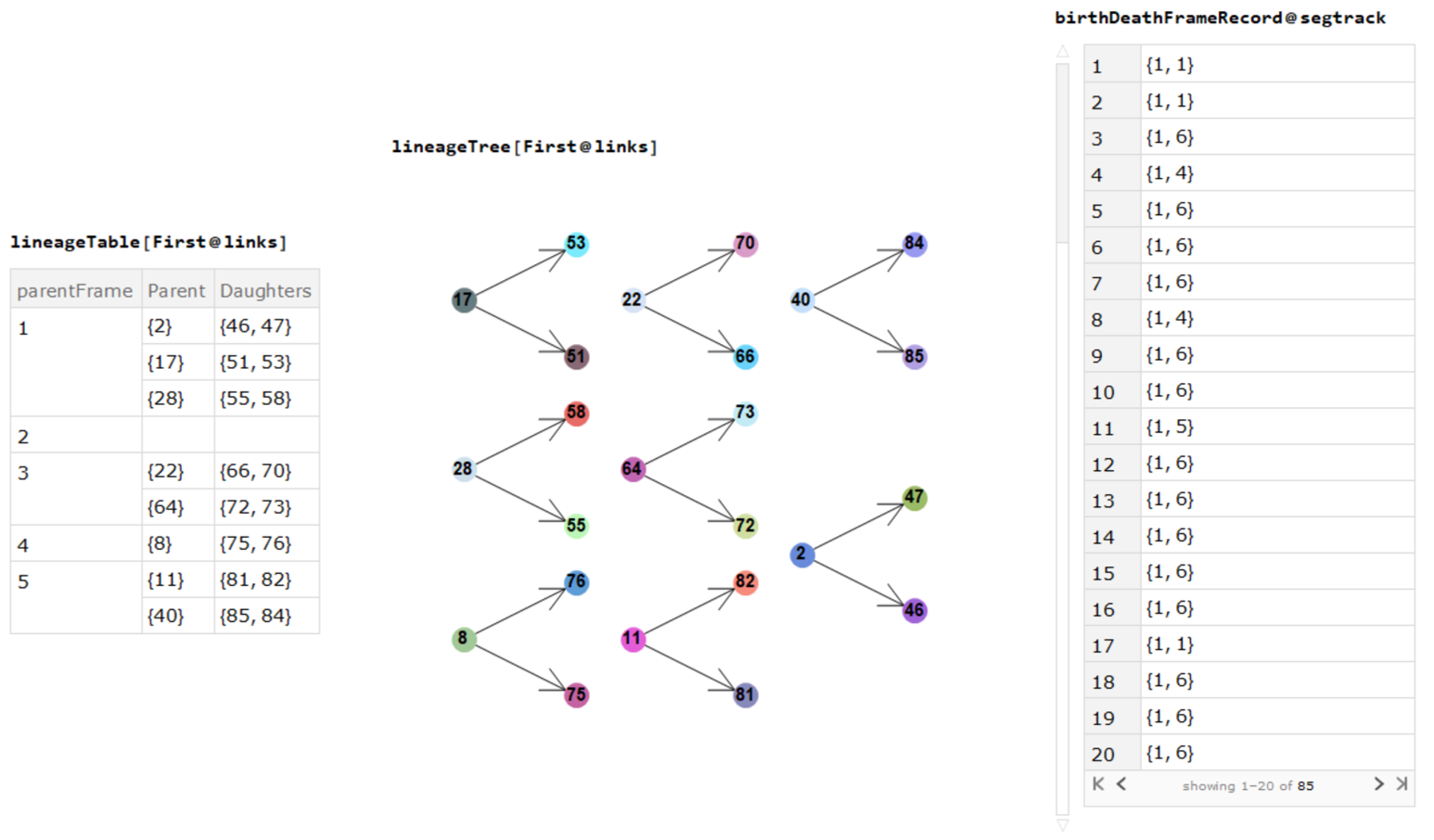
division events, mother-daughters cell indices and cell births/deaths
I hope that future versions of Mathematica can incorporate more from the areas of image processing and computer vision
CODE
LineageMapper
BeginPackage["LineageMapper`",{"segmentStack`"}];
cellTracker::usage = "cellTracker implements an overlap-based algorithm for tracking cells in timelapse images. The algorithm is able to robustly track cell movements
across frames independently of the segmentation scheme employed, detects mitotic events and corrects any incorrect mergers of cells in confluent conditions. Both binarized masks
or labelled matrices can be provided to this particular implementation. The output is in the form of relabelled matrices identifying the cells with unique labels.
Note: the algorithm is based on the approach proposed by Chalfoun et al. Scientific Reports, 2016.";
(* params *)
Options[cellTracker] = {"segmented" -> False, "overlapW" -> 1, "sizeW" -> 0.2, "centroidW" -> 0.5, "maxCentDist" -> 50.0, "mincellsize" -> 100,
"criteria"-> {"on","on"}, "pdthreshoverlap" -> 0.2, "sizeSimilarity" -> 0.5, "aspectSimilarity" -> 0.7, "parentCircularity" -> 0.3,
"frameCircularitycheck" -> 5, "fusionOverlapThresh" -> 0.2,"FusionEnabled" -> False};
(* Overlap Matrix *)
(*
(* overlap matrix between the frames *)
overlapMatrix[segPrev_,segCurr_]:=Module[{labelPrev, maxLabelCurr,labelCurr},
labelPrev = Values@ComponentMeasurements[segPrev,"Label"];
labelCurr = ComponentMeasurements[segCurr,"Label"];
maxLabelCurr = Max@Keys@labelCurr;
Normal@SparseArray[
SparseArray[#,maxLabelCurr]&@
DeleteCases[Normal@*Counts@Flatten[(1-Unitize[segPrev-#])segCurr], 0 -> _]&/@labelPrev,
{Length@labelPrev,Length@labelCurr}
]
];
*)
(* this scheme is faster than the one above *)
overlapMatrix[seg1_,seg2_]:=Block[{keys1,keys2,mask,map,rules1,rules2},
keys1 = Keys@ComponentMeasurements[seg1, "Label"];
keys2 = Keys@ComponentMeasurements[seg2, "Label"];
mask= Unitize[seg1*seg2];
map = Normal@Counts@Thread[{SparseArray[mask*seg1]["NonzeroValues"],SparseArray[mask*seg2]["NonzeroValues"]}];
{rules1,rules2}= Dispatch@Thread[# -> Range@Length@#]&/@{keys1,keys2};
map[[All,1,1]]=map[[All,1,1]]/.rules1;
map[[All,1,2]]=map[[All,1,2]]/.rules2;
Normal@SparseArray[map,{Length@keys1,Length@keys2}]
];
(* Cost Matrix *)
(* for determining the overlapTerms *)
overlapCompiled = Compile[{{overlapmat, _Integer, 2},{prevList, _Real, 1},{currList, _Real, 1}},
1 - (overlapmat/(2.0 prevList) + (overlapmat\[Transpose] /(2.0 currList))\[Transpose]),
CompilationTarget -> "C"
];
(* for determining the sizeTerm *)
sizeCompiled[prevList_, currList_] := With[{curr = currList},
Abs[# - curr]/Clip[curr,{#,\[Infinity]}]&/@prevList
];
(* sizeCompiled = Compile[{{prevList,_Real,1},{currList,_Real,1}},
Outer[Abs[#1-#2]/Max[#1,#2]&, prevList, currList],
CompilationTarget \[Rule] "C"]; *)
(*for determining the centroidTerm *)
centCompiled = Compile[{{centDiffMat, _Real, 2},{threshold, _Real}},
Map[If[# >= threshold, 1., #/threshold]&, centDiffMat,{2}],
CompilationTarget-> "C"
];
(* centCompiled = Compile[{{centDiffMat, _Real, 2}, {threshold, _Real}},
threshold UnitStep[centDiffMat - threshold] + (1 - UnitStep[centDiffMat - threshold]) centDiffMat,
CompilationTarget -> "C"] (* needs to be modified for #/threshold (see above) *)
*)
(* as the name implies, costMatrix generates the cost of traversing between an object @t and @t+1. More metrics e.g. texture metrics can be added.
In fact an arbitrary # of user defined metrics can be incorporated to compute the cost *)
costMatrix[segPrev_,segCurr_,overlapMat_, OptionsPattern[cellTracker]]:= Module[{centroidPrev,centroidCurr, centroidDiffMat,
areaPrev,areaCurr, nCol,nRow, overlapTerm,costMat,pos,centroidTerm,sizeTerm,spArrayOverlap,spArraycentDiff,mask,
overlapW = OptionValue@"overlapW", sizeW = OptionValue@"sizeW", centroidW = OptionValue@"centroidW",
maxCentDist = OptionValue@"maxCentDist"},
{centroidPrev,areaPrev} = Values@ComponentMeasurements[segPrev,{"Centroid","Area"}]\[Transpose];
{centroidCurr,areaCurr} = Values@ComponentMeasurements[segCurr,{"Centroid","Area"}]\[Transpose];
{nRow,nCol} = {Length@areaPrev,Length@areaCurr};
centroidDiffMat = DistanceMatrix[N@centroidPrev,N@centroidCurr]; (* calculating pairwise distance between centroids in frames t, t+1 *)
centroidTerm = centCompiled[centroidDiffMat,maxCentDist]; (* optimized to compute centroidTerm *)
overlapTerm = overlapCompiled[overlapMat,areaPrev,areaCurr]; (* optimized for speed to compute overlapTerm *)
sizeTerm = sizeCompiled[areaPrev,areaCurr]; (* optimized for speed to compute sizeTerm *)
spArrayOverlap = SparseArray[overlapTerm, Automatic, 1.]; (* finding positions in overlapTerm where cells intersect *)
spArraycentDiff = SparseArray[UnitStep[maxCentDist - centroidDiffMat], Automatic, 0]; (* finding positions in centDiffMat for dist within maxCentDist *)
pos = spArrayOverlap["NonzeroPositions"]~Union~spArraycentDiff["NonzeroPositions"]; (* positions where overlaps occur or centroids within maxCentDist *)
mask = SparseArray[pos->1,{nRow,nCol},\[Infinity]]; (* creating the mask for costMat *)
mask*(overlapW*overlapTerm + centroidW*centroidTerm + sizeW*sizeTerm)
];
(* Row/Column minimization schemes *)
(* row wise minimums to determine which target cells are mapped from the source cells (previous frame) *)
rowwiseMins[costMat_]:=With[{constInfArray = ConstantArray[\[Infinity],Last@Dimensions[costMat]]},
Rule@@@SparseArray[Unitize@Map[If[Min[#]==\[Infinity], constInfArray, # - Min@#]&,costMat], Automatic, 1]["NonzeroPositions"]
];
(* column wise minimums to determine cell mappings from current frame to the previous frame *)
colwiseMins[costMat_,groupingmetric_:(Last->First)]:= With[{constInfArray = ConstantArray[Infinity,First@Dimensions[costMat]]},
GroupBy[
SparseArray[Unitize@Map[If[Min[#]==Infinity,constInfArray ,# - Min[#]]& , costMat\[Transpose]], Automatic, 1]["NonzeroPositions"],
groupingmetric,#]&
];
(* Mitosis Check *)
(* subF[] with its HoldFirst attribute creates isTrueDivision[] to check whether the daughters meet the criteria *)SetAttributes[subF,HoldFirst];
subF[body_]:= Block[{seg,parent,daughterpair},
With[{$boundingCurr = Unevaluated@Values@ComponentMeasurements[seg, "SemiAxes"]},
SetDelayed@@(Hold[isTrueDivision[seg_][parent_, daughterpair_],
With[{aspectRatioN = Composition[Max[#]/Min[#] &, Identity] /@ $boundingCurr},
With[{aspectr = aspectRatioN[[daughterpair]]},
body]
]
] /. Unevaluated -> Sequence)
]
];
(* 1st option \[Rule] size comparision, 2nd option \[Rule] aspectratio comparison *)
initializeF[OptionsPattern@cellTracker] := Block[{parent, daughterpair, seg, aspectr$},
With[{crit = OptionValue["criteria"], sizeSim = OptionValue["sizeSimilarity"], aspectSim = OptionValue["aspectSimilarity"],
areas = Unevaluated@(Values[ComponentMeasurements[seg, "Area"]][[daughterpair]])},
Switch[crit,
{"off","off"},
SetDelayed@@(Hold[isTrueDivision[seg_][parent_, daughterpair_], True]),
{"on", "off"},
SetDelayed@@(Hold[isTrueDivision[seg_][parent_, daughterpair_], (1 - Abs[Subtract@@areas/Total@areas] >= sizeSim)] /. Unevaluated -> Sequence),
{"off", "on"},
subF[(1 - Abs[Subtract@@aspectr$/Total@aspectr$] >= aspectSim)],
{"on", "on"},
subF[(1 - Abs[Subtract@@areas/Total@areas] >= sizeSim) && (1 - Abs[Subtract@@aspectr$/Total@aspectr$] >= aspectSim)]
]
]
];
(* checkDivision[ ] yields any mitotic events if they pass the stringent criterion *)
checkDivision[costMat_,overlapMat_,segCurr_,segPrev_, trackvector_, colwisemap_,OptionsPattern@cellTracker]:= With[{pdthreshoverlap = OptionValue@"pdthreshoverlap",
circularityframeCheck = OptionValue@"frameCircularitycheck",parentcircThresh = OptionValue@"parentCircularity"},
Module[{potentialparentstodaughter,
potentialDPoverlap,indices, multipledaughterspairs, parents, selcosts, getcosts,selpos,parentsKeys,selpairsAssociation,PtoFmapping,FtoPmapping,
multimappings,mothercircularity,combinedFrameHist, areaCurr,areaPrev, isTrueDivisionQ,picks,daughtermappingOtherCells,mappeddaughters,trueElems},
areaCurr = Values@ComponentMeasurements[segCurr,"Area"];
areaPrev = Values@ComponentMeasurements[segPrev,"Area"];
(* if all cells @ t+1 map to a single cell @ t then no division events *)
If[AllTrue[Values@colwisemap[Length],# == 1 &], Return[{}]];
(* execution got here, means there are potential division events *)
potentialparentstodaughter = Flatten@Cases[Normal@colwisemap[Identity],HoldPattern[_ -> {Repeated[_Integer,{2,\[Infinity]}]}]];
(* do the potential daughter-parents overlap significantly *)
potentialDPoverlap = KeyValueMap[#1-> overlapMat[[##]]/areaCurr[[#2]]&,<|potentialparentstodaughter|>];
potentialparentstodaughter = Pick[potentialparentstodaughter,potentialDPoverlap, x_/;x >= pdthreshoverlap]/.
HoldPattern[x_Integer -> {}] :> Sequence[];
(* remove potential daughters that are tracked to cells other cell, other than potential parent, if they overlap with the
cells they are tracked to *)
PtoFmapping=Reverse[trackvector,{2}];
FtoPmapping =Association@@@Map[Reverse[#,{2}]&@*Thread]@potentialparentstodaughter;
parents = Keys@potentialparentstodaughter;
multimappings= Normal@Map[(
Select[
KeySelect[GroupBy[Join[Normal@#,PtoFmapping],First-> Last,Union]
,<|Thread[Keys[#]-> True]|>],
Length@#>1&])&,FtoPmapping];
daughtermappingOtherCells = MapThread[Replace[#,{#2-> Sequence[]},{3}]&,{multimappings,parents}];
mappeddaughters = Replace[daughtermappingOtherCells,{HoldPattern[x_ -> _]}:>x, \[Infinity]];
indices = Replace[Reverse[daughtermappingOtherCells,{3}],{p:HoldPattern[{__Integer}-> _]}:> List@@@Thread@p,{1}];
trueElems=Map[overlapMat[[Sequence@@#]] > 0 &,indices, {2}]/.{True..}:> True;
potentialparentstodaughter = MapThread[If[#1 === True,
First@#2->Complement[Last@#2,{#3}],#2]&,{trueElems,potentialparentstodaughter,mappeddaughters}];
(* if potential daughters are 0 or 1 then no division event for the parent and hence remove *)
potentialparentstodaughter = potentialparentstodaughter /. HoldPattern[_ -> {}|{_}]:> Sequence[];
(* for potential parents with > 2 daughters take the pair with the lowest costs *)
multipledaughterspairs = <|Cases[potentialparentstodaughter, HoldPattern[_ -> {Repeated[_Integer,{3,\[Infinity]}]}]]|>;
parents = Keys@multipledaughterspairs;
getcosts = KeyValueMap[costMat[[##]]&,multipledaughterspairs];
selcosts = TakeSmallest[#,2]&/@getcosts;
selpos = Position[getcosts,Alternatives@@Flatten@selcosts];
parentsKeys = Part[parents,selpos[[All,1]]];
selpairsAssociation = GroupBy[MapThread[#1-> multipledaughterspairs[[Sequence@@#2]]&,{parentsKeys,selpos}], First -> Last];
potentialparentstodaughter =Normal@SortBy[Join[<|potentialparentstodaughter|>,selpairsAssociation],First];
(* only select pairs wherein both daughters overlap with their parent significantly *)
potentialDPoverlap=KeyValueMap[Total@(overlapMat[[##]]/Part[areaPrev,#1])&,<|potentialparentstodaughter|>];
potentialparentstodaughter=Pick[potentialparentstodaughter,potentialDPoverlap,x_/;x >= pdthreshoverlap];
(* circularity check for mothers: if $framenumber is < frameCircularitycheck, then set all the parentscircularity to True *)
mothercircularity = If[Length@$framehistory <= circularityframeCheck,
Array[True&,Length@*Keys@potentialparentstodaughter],
combinedFrameHist=GroupBy[Flatten@$framehistory,First-> Last];
Replace[Map[combinedFrameHist,parents],{x_/;Length@x<=circularityframeCheck-> True,
x_/;AnyTrue[x, # >= parentcircThresh &]-> True,_ -> False},{1}]
];
potentialparentstodaughter=Pick[potentialparentstodaughter,mothercircularity,True];
(* daughters should meet the aspect and size similarity criteria *)
isTrueDivisionQ = isTrueDivision[segCurr];
picks = Map[isTrueDivisionQ@@#&,potentialparentstodaughter];
Extract[potentialparentstodaughter,Position[picks,True]]/.Rule-> List
]
]
(* Fusion Check *)
(* identifyFusions[] gives the possible indices of cells @t that merged @t+1. *)
identifyFusions[currSeg_,prevSeg_,overlapMat_,trackvector_,OptionsPattern@cellTracker]:= With[{fusionOverlapThresh = OptionValue["fusionOverlapThresh"]},
Module[{fmatrix, fvector,
nrow, ncol, nullcandidates, areaprev= Values@ComponentMeasurements[prevSeg,"Area"], indices},
nrow = Length@areaprev;
ncol = Length@ComponentMeasurements[currSeg,"LabelCount"];
fmatrix = SparseArray[(trackvector /. Rule -> List) -> 1, {nrow,ncol}];
fvector = Total@fmatrix;
nullcandidates = Flatten@Position[fvector,Except[0,x_/;x<2], Heads->False];
fmatrix[[All,nullcandidates]] = ConstantArray[0,nrow];
indices = Map[Reverse]@(Replace[(fmatrix*UnitStep[(overlapMat/areaprev) - fusionOverlapThresh])["NonzeroPositions"],
p:{{__Integer}}:> Flatten@p]); (* fusion cases *)
Normal@GroupBy[
Cases[GatherBy[indices, First],{p:Repeated[{_,_},{2,\[Infinity]}]}:> p],
First->Last] (* we can also use Gather[indices, First@#2 \[Equal] First@# &] instead of GatherBy *)
]
]
(* breakingFusion will break the blob/cluster/incorrect fusion of cells @t+1 into the individual cells, that they were @ t*)
breakingFusion[{currSeg_,daughters_},prevSeg_,parentkeys_,OptionsPattern@cellTracker][rule_]:= Block[{newLabels,
blobind,cluster,tempSeg,unassignedpos,t,$i=1,nf,replaceResidual,blobpos,masks, boundmasks,intersectionpos, symindices,
keys, rules, newlabelblob, keyscurr,conflicts,replaceRulesCurr, toreplace, confdaughters, symDs, newDaughterLab,
oldDaughterLab, DaughterLabChange = {}},
keyscurr = Keys@ComponentMeasurements[currSeg,"Label"]; (* labels of cells in current frame *)
newlabelblob = Max@parentkeys + Max@keyscurr + Length@Last@rule + 1; (* possible new label for the blob in current frame *)
{blobind,cluster} = rule /. Rule -> List; (* {blob,{cluster}} *)
(* is daughter(s) part of conflicts: assign a different label @ the end if so *)
confdaughters = daughters \[Intersection] cluster;
oldDaughterLab = Extract[daughters, Position[daughters, Alternatives@@confdaughters]];
(* get potential conflicts between 't' and 't+1' frame *)
conflicts = keyscurr \[Intersection] cluster;
symindices = Table[Unique[], Length@conflicts];
(* symbols for conflicting daughters *)
symDs = Extract[symindices,Position[conflicts,Alternatives@@confdaughters]];
(* change blob index to the new index i.e. newlabelblob and conflicts to syms *)
replaceRulesCurr = {Thread[blobind -> newlabelblob]}~Join~Thread[conflicts -> symindices];
tempSeg = Replace[currSeg,replaceRulesCurr,{2}];
blobpos = SparseArray[Unitize[newlabelblob - tempSeg]/. _Unitize -> 1,Automatic, 1]["NonzeroPositions"]; (* position of blob in current frame *)
tempSeg = Block[{mask, modifiedseg, row = First@Dimensions[prevSeg], col = Last@Dimensions[currSeg]},
Fold[(intersectionpos = blobpos \[Intersection] SparseArray[Unitize[#2 - prevSeg], Automatic, 1]["NonzeroPositions"];
mask = SparseArray[(t[$i++] = intersectionpos) -> 1, {row,col}];
#2 mask + (1 - mask) #)&, tempSeg, cluster]
];
unassignedpos = SparseArray[Unitize[newlabelblob - tempSeg]/._Unitize -> 1,Automatic,1]["NonzeroPositions"]; (* residual part of blob *)
nf = Nearest@Flatten@MapThread[Thread[#1->#2]&,{Array[t,$i-1],cluster}];
replaceResidual = Replace[Map[#->nf[#,1]&, unassignedpos], HoldPattern[p:{_,_}-> {x_}]:> p -> x,{1}]; (* assign nearest label to residual position *)
tempSeg = ReplacePart[tempSeg,replaceResidual]; (* replace residual *)
(* rename symbols to integer labels *)
If[conflicts != {},
keys = DeleteCases[Keys@*Counts@*Flatten@tempSeg, 0];
newLabels = Complement[Range[Length@keys], keys];
toreplace = Cases[keys,_Symbol];
rules = Thread[toreplace -> Take[newLabels,Length@toreplace]];
If[symDs != {},
newDaughterLab = Cases[rules,PatternSequence[Alternatives@@symDs -> ind_]:> ind];
DaughterLabChange = Thread[oldDaughterLab -> newDaughterLab];
];
tempSeg = Replace[tempSeg,rules,{2}]
];
masks = Cases[Last@@@ComponentMeasurements[tempSeg,{"Label","Mask"},
MemberQ[Alternatives@@cluster,#]&], _SparseArray, {2}]; (* position of new labels in "Label" *)
boundmasks = Map[ColorNegate@*MorphologicalPerimeter@*Image, masks]; (* boundary mask of the newly split cells *)
tempSeg = Fold[# ImageData[#2]&, tempSeg, boundmasks]; (* create distinct boundaries for the split cells *)
(* delete cells smaller than the threshold \[Rule] needs to be checked *)
{Fold[Block[{unitize = Unitize[#2-#1]},
If[Length@SparseArray[unitize, Automatic, 1]["NonzeroValues"] <= OptionValue["mincellsize"],
unitize #1, #1]]&,tempSeg,cluster], daughters /. DaughterLabChange}
];
(* The Assignment Problem *)
(* the function operates on the cost matrix etc.. to create a mapping between the source and target cells if any. Any unassigned cells @t are either
dead or mitotic mother cells. Similarly any unassigned cells @t+1 either enter the FOV or are daughters. Fused cells once broken maintain their original
labels. Here we use a graph with edgeweights to find unique mappings *)
assignmentFunc[segCurr_,trackvector_,costMat_,fusionpairs_,mdpairs_,truePrevKeys_,opt:OptionsPattern[cellTracker]]:= With[{fusionflag = OptionValue["FusionEnabled"]},
Module[{segmentCurr= segCurr,
currframelabels, currlabelsUntracked, daughters, mothers, realindices,artificialInds,newlabels,assignmentsList,
ruleAssigned, currentassigned, currentunassigned, newcells, maxlabelprev,newcellAssignmentRules,allAssignmentRules,
daughterlabels,previnds,selfRules,otherassoc,rules,fusionsP,fusionsQ,fusedlabels},
currframelabels = Keys@ComponentMeasurements[segCurr,"Label"];
{fusionsQ,fusionsP} = {Keys@#,Values@#}&[fusionpairs];
{mothers,daughters}= mdpairs /.{{}-> {{},{}},x_/;True :> Thread@x};
daughters = Flatten@daughters;
currlabelsUntracked = Complement[currframelabels, Join[daughters,Flatten@fusionsP]];
(* other possible associations using columnwise mins *)
otherassoc= Sort@Flatten@KeyValueMap[Thread@*Rule,DeleteCases[colwiseMins[costMat]@Identity,x_/;Length@x>1]];
rules=Sort@DeleteDuplicates[Join[trackvector,otherassoc]];
artificialInds = rules/.Rule -> List;
previnds = Part[truePrevKeys,artificialInds[[All,1]] ];
realindices = Transpose[{previnds,Part[artificialInds,All,2]}];
(* create the graph with initialized weights for the assignment problem *)
assignmentsList = Block[{p,c,edges,edgeweights,graph,assignments},
edges=Subscript[p, #[[1]]]-> Subscript[c, #[[2]]]&/@realindices;
edgeweights = costMat[[Sequence@@#]]&/@artificialInds;
graph = Graph[edges,EdgeWeight->1./edgeweights];
assignments= FindIndependentEdgeSet@graph;
Replace[assignments,HoldPattern[Subscript[p, x_]\[DirectedEdge]Subscript[c, y_]] :> {x,y},{1}]
];
ruleAssigned = Reverse[Rule@@@assignmentsList,{2}];
currentassigned = Part[assignmentsList,All,2];
currentunassigned = Complement[currlabelsUntracked,currentassigned]; (* these are cells that arrived into FOV *)
newcells = If[!fusionflag,Sort@#, Sort[#~Join~fusionsP]]&@Catenate[{currentunassigned,daughters}];
maxlabelprev=Max@truePrevKeys;
newlabels=Range[maxlabelprev+1,maxlabelprev+Length@newcells];
newcellAssignmentRules=Thread[newcells-> newlabels];
selfRules = If[!fusionflag, fusionsP/.{{}-> {},x_/;True:>(Thread[#-> #]&@Flatten@x)},{}];
allAssignmentRules = Dispatch@SortBy[newcellAssignmentRules~Join~ruleAssigned~Join~selfRules,First];
(* sow true labels of mother with the new labels of daughters *)
mothers = Part[truePrevKeys,mothers];
daughterlabels = Partition[Lookup[<|newcellAssignmentRules|>,daughters],2];
If[Not@fusionflag, Sow@<|Thread[mothers -> daughterlabels]|>,
((*get new labels for the fusion case and sow if fusion is enabled *)
fusedlabels = Lookup[<|newcellAssignmentRules|>,fusionsP];
{Sow[<|Thread[mothers -> daughterlabels]|>,"x"], Sow[<|Thread[fusedlabels -> fusionsQ]|>,"y"]}
)
];
Replace[segCurr,allAssignmentRules,{2}] (* Use replace instead of arraycomponents with dispatch *)
]
]
(* Mains *)
SetAttributes[updateMat, HoldFirst];
updateMat[mat_,r_,c_,opt:"cost"|"overlap"]/;r!={}:=mat=Switch[opt,"cost", (mat[[r,All]]= \[Infinity];
mat[[All,Flatten@c]]= \[Infinity]; mat), "overlap", (mat[[r,All]]= 0;
mat[[All,Flatten@c]]= 0; mat)]
relabelMat[segmentCurr_,segmentPrev_,mothersFindex_,daughters_, splits_]:= Module[{segCurr = segmentCurr,
indicesTokeep, keeplabelRules,overlaps, costMat},
indicesTokeep = daughters~Join~splits;
keeplabelRules = Flatten@Map[#->#&,indicesTokeep,{2}];
segCurr = ArrayComponents[segmentCurr,2,Sort@keeplabelRules];
overlaps = overlapMatrix[segmentPrev,segmentCurr]; (* recomputing overlap and cost-matrix *)
costMat = costMatrix[segmentPrev,segmentCurr,overlaps];
updateMat[overlaps,mothersFindex,daughters,"overlap"]; (* modifying overlap matrix in place *)
updateMat[costMat,mothersFindex,daughters,"cost"]; (* modifying cost matrix in place *)
(* updateMat[#1,mothersFindex,daughters,#2]&@@@{{costMat,"cost"},{overlaps,"overlaps"}}; *)
{costMat,overlaps,segCurr,rowwiseMins@costMat}
]
(* a helper function to the Mains to run the algorithm *)
stackCorrespondence[segmentPrev_, Current_, opt:OptionsPattern[cellTracker]]:= With[{frameCircCheck = OptionValue["frameCircularitycheck"],
segmentedQ = OptionValue["segmented"],fusionflag = OptionValue["FusionEnabled"]},
Module[{overlaps,segmentCurr,costMat,daughters,curr = Current,trackvector,colwisemap,truePrevkeys,mothersFindex,
fusionsFindex,fusionsTindex, mdpairs,splits,indicestokeep,keeplabelsRules,flattenDaughters, Dlabelchanges,cluster},
segmentCurr = Switch[segmentedQ, False, segmentImage@Import@curr, _, curr]; (* segmentation ? \[Rule] do or do not as Yoda says *)
overlaps = overlapMatrix[segmentPrev,segmentCurr]; (* compute overlaps *)
costMat = costMatrix[segmentPrev,segmentCurr,overlaps]; (* associated costMatrix between the two segmented images *)
truePrevkeys = Keys@ComponentMeasurements[segmentPrev,"Label"]; (* actual labels for parents @ t *)
(* computing column mins and row mins *)
{trackvector,colwisemap} = Through[{rowwiseMins,colwiseMins}[costMat]];
(* history about parent's roundness *)
If[Length@$framehistory <= frameCircCheck,
AppendTo[$framehistory,
MapAt[4 \[Pi] (First[#]/(Last[#]^2))&, ComponentMeasurements[segmentPrev,{"Area","PerimeterLength"}], {All, 2}]],
$framehistory = Delete[$framehistory,1]
];
(* appropriate checks for mitotic events *)
{mothersFindex,daughters} = (checkDivision[costMat,overlaps,segmentCurr,segmentPrev,trackvector,colwisemap])/.{(_Return -> {{},{}}),
(x_/; x =!={} :> Transpose[x]), _?(#=={}&) :> {{},{}}};
mdpairs = Thread[{mothersFindex,daughters}];
flattenDaughters = Flatten@daughters;
(* if mitotic events exist then replace the rows for the costmatrix and the columns (daughters) with \[Infinity] and for overlap matrix with 0.
This removes all possible paths from parents to cells @t+1 and for cells @t to reach daughters *)
updateMat[costMat,mothersFindex,daughters,"cost"]; (* modifying cost matrix in place *)
updateMat[overlaps,mothersFindex,daughters,"overlap"]; (* modifying overlap matrix in place *)
(* updateMat[#1,mothersFindex,daughters,#2]&@@@{{costMat,"cost"},{overlaps,"overlaps"}}; *)
trackvector = rowwiseMins@costMat; (* update tracking vector between frames *)
(* identify fusions events *)
fusionsFindex = identifyFusions[segmentCurr,segmentPrev,overlaps,trackvector];
fusionsTindex = Map[Part[truePrevkeys,#]&, fusionsFindex,{3}];
Which[Not@fusionflag,
(* breaking incorrectly fused clusters. this will create new cells in the current frame. the cost matrix and overlap matrix need to be recomputed *)
(If[fusionsTindex != {},
{segmentCurr,flattenDaughters} = Fold[breakingFusion[#,segmentPrev,truePrevkeys][#2]&,{segmentCurr,flattenDaughters},fusionsTindex];
If[flattenDaughters != {},
daughters = Partition[flattenDaughters,2];
mdpairs = Thread[{mothersFindex,daughters}]
];
];
If[fusionsTindex != {} || mothersFindex != {},
splits = Values@fusionsTindex;
indicestokeep = daughters~Join~splits;
keeplabelsRules = Flatten@Map[#->#&,indicestokeep,{2}];
segmentCurr = ArrayComponents[segmentCurr,2,Sort@keeplabelsRules];
overlaps = overlapMatrix[segmentPrev,segmentCurr]; (* recomputing overlap and cost-matrix *)
costMat = costMatrix[segmentPrev,segmentCurr,overlaps];
updateMat[overlaps,mothersFindex,daughters,"overlap"]; (* modifying overlap matrix in place *)
updateMat[costMat,mothersFindex,daughters,"cost"]; (* modifying cost matrix in place *)
trackvector = rowwiseMins@costMat (* recomputing trackvector *)
]; (* assignments are made *)
assignmentFunc[segmentCurr,trackvector,costMat,fusionsTindex,mdpairs,truePrevkeys,opt]),
True (* when fusion is enabled *),
(* Modify cost/overlap matrix if possible and assign directly since we already know the cluster label(s) if any
and parent/daughter label(s) if any *)
cluster = Keys@fusionsTindex;
updateMat[overlaps,mothersFindex,daughters~Join~cluster,"overlap"]; (* modifying overlap matrix in place *)
updateMat[costMat,mothersFindex,daughters~Join~cluster,"cost"]; (* modifying cost matrix in place *)
trackvector = rowwiseMins@costMat; (* recomputing trackvector *)
(* make assignments *)
assignmentFunc[segmentCurr,trackvector,costMat,Reverse[fusionsTindex,2],mdpairs,truePrevkeys,opt]
]
]
];
(* Mains Function *)
cellTracker[files_, opt: OptionsPattern[]]:= Module[{option = OptionValue["segmented"], Prev},
initializeF[]; (* initialize *)
Prev = Switch[option, False, segmentImage@*Import@*First@files, _, First@files];
Reap@FoldList[stackCorrespondence[##,opt]&, Prev, Rest@files]
];
End Package [ ]
Begin["`Private`"];
$framehistory = {};
End[];
EndPackage[];
 Attachments:
Attachments: