Introduction
Malaria is a life-threatening disease caused by parasites that are transmitted to people through the bite of infected female Anopheles mosquitoes. Once inside the body, the Malaria parasite multiplies in the liver cells and gets released back into the bloodstream to destroy blood cells. In 2017, there were an estimated 219 million cases of Malaria in 87 countries. The symptoms of malaria include cycles of chills, fever, sweats, muscle aches and headache that recur every few days. There can also be vomiting, diarrhea, coughing, and jaundice in the skin and eyes. Even more extreme symptoms include, bleeding problems, shock, kidney and liver failure, central nervous system problems, comas, and death. A diagnosis is When detected early and accurately, such symptoms can be prevented, thus using machine learning to quickly, efficiently, and reliably diagnose malaria is important to help people stop the negative effects of malaria before its too late.
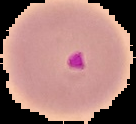
Below is an example of a Malaria Uninfected Cell compared to a Malaria Infected Cell:

Data Importing
I imported a data set of malaria infected and uninfected red blood cells from Kaggle. Each class was placed in its corresponding variable.
infected =
FileNames["*.png",
"C:\\Users\\Shruti Panse\\Desktop\\Shruti Panse- Wolfram Summer \
Camp\\WSS-Template\\Final Project\\Drafts\\cell_images\\Parasitized"];
uninfected =
FileNames["*.png",
"C:\\Users\\Shruti Panse\\Desktop\\Shruti Panse- Wolfram Summer \
Camp\\WSS-Template\\Final Project\\Drafts\\cell_images\\Uninfected"];
Constructing File Objects for Images
I needed to match each blood cell image with a value of either true for infected or false for uninfected. In order to increase the efficiency of the importation, I created separate file objects for each of the image variables. Each variable contained 13,779 images for parisitized and uninfected cells.
infectedIMG = File /@ infected;
uninfectedIMG = File /@ uninfected;
Next, I made a list of 13,779 true and false values which would later be used to be matched up with their respective images. I placed these lists in variables, and made another variable to connect the list of true and false values together along with another variable that connected the infected and uninfected file objects together.
Length[infected]
13779
infectedvalues = Table[True, Length[infected]];
Length[uninfected]
13779
uninfectedvalues = Table[False, Length[uninfected]];
Finally, using the AssociationThread function, I linked the images with their values and separated the data into two groups, 75% for training and 25% for validation.
data = RandomSample[AssociationThread[imagekeys -> values]];
traininglength = Length[data]*.75
20668.5
trainingdata = data[[1 ;; 20669]];
validationdata = data[[20670 ;;]];
Creating the Neural Network
Next, it was time to start on the Neural Network, which used MNIST image classification as the basis for its structure. The network's purpose was to classify uninfected and infected cells using true and false to describe the type of cell. I created a NetChain function that had many layers. One notable layer is the Resize layer which is used to change the image dimensions of each image to 135 by 135. This adjusts the images to comply with the neural networks sensitivity to the size of images. Further layers included the convolution layer, ramp, and pooling layer which all worked to narrowed down features and create classes that to identify each image to compare them.
dims = {135, 135}
{135, 135}
lenet = NetChain[{
ResizeLayer[dims],
ConvolutionLayer[20, 5],
Ramp, (*Takes out the the not useful features*)
PoolingLayer[2, 2],(*Downsamples*)
ConvolutionLayer[50, 5],
Ramp, (*Takes out the the not useful features*)
PoolingLayer[2, 2],(*Downsamples*)
FlattenLayer[], 500,(*Makes features into feature vector"*)
Ramp, 2,(*Takes out the the not useful features- True or false*)
SoftmaxLayer[]},(*Turns the vector into probabilities*)
"Output" ->
NetDecoder[{"Class", {True, False}}],(*Tensor into true or false*)
"Input" -> NetEncoder["Image"](*Turns image into numbers*)
]
Training the Neural Networks with NetTrain
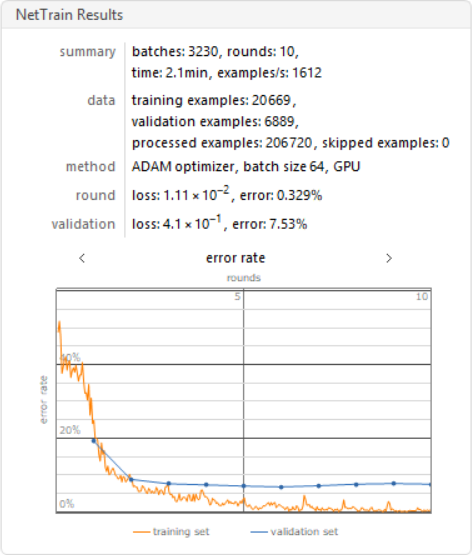
I trained the neural nets with 10 training rounds on a GPU.
results =
NetTrain[lenet, Normal[trainingdata], All,
ValidationSet -> Normal[validationdata], MaxTrainingRounds -> 10,
TargetDevice -> "GPU"]
Training the Neural Network with Augmented Layers
Next I implemented an ImageAugmentationLayer, which randomly crops images to create new data sets to improve my neural network.
augment = ImageAugmentationLayer[{135, 135},
"Input" -> NetEncoder[{"Image", {139, 139}}],
"Output" -> NetDecoder["Image"]]
I made the images 139 by 139 and allowed the augmentation layer to crop the images by 4 pixels at random within the constraints of the dimensions of 135 by 135.
dims2 = {139, 139}
lenet2 = NetChain[{
ResizeLayer[dims2],
ImageAugmentationLayer[{135, 135}],
ConvolutionLayer[20, 5],
Ramp,
PoolingLayer[2, 2],
ConvolutionLayer[50, 5],
Ramp,
PoolingLayer[2, 2],
FlattenLayer[], 500,
Ramp, 2,
SoftmaxLayer[]},
"Output" -> NetDecoder[{"Class", {True, False}}],
"Input" -> NetEncoder["Image"]
]
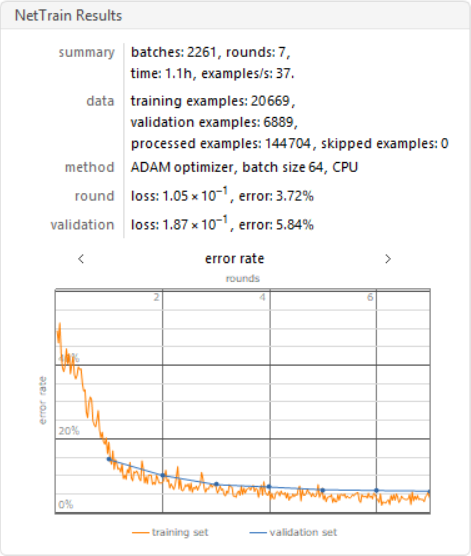
I trained this data using the neural net, with only 7 layers and on a CPU.
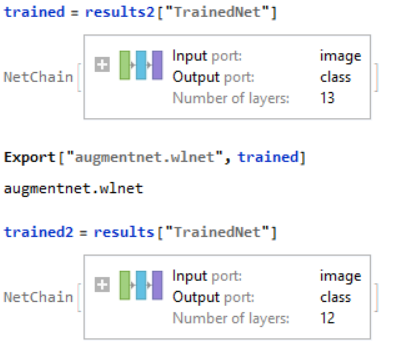
results2 =
NetTrain[lenet2, Normal[trainingdata], All,
ValidationSet -> Normal[validationdata], MaxTrainingRounds -> 7]
Creating a Testing Set for Data
Data Visualization
Lastly, I made a ConfusionMatrixPlot using the Classifier Measurements function which compares the neural networks predicted class against the actual class result.
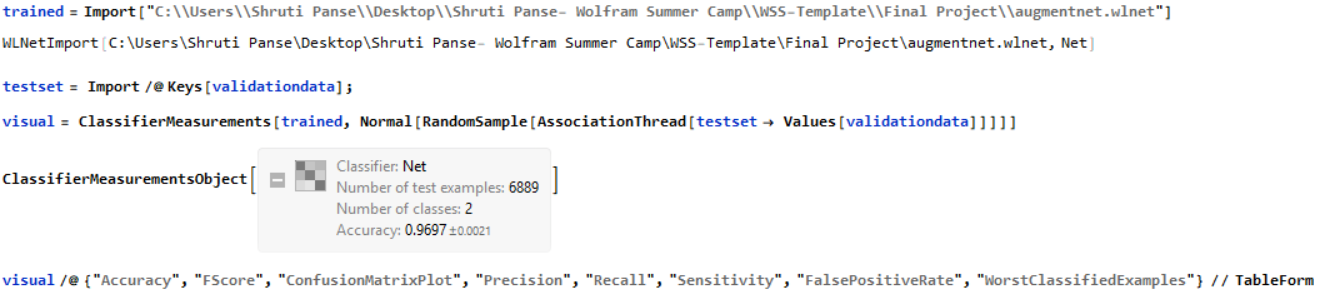 Below is the Matrix:
Below is the Matrix: 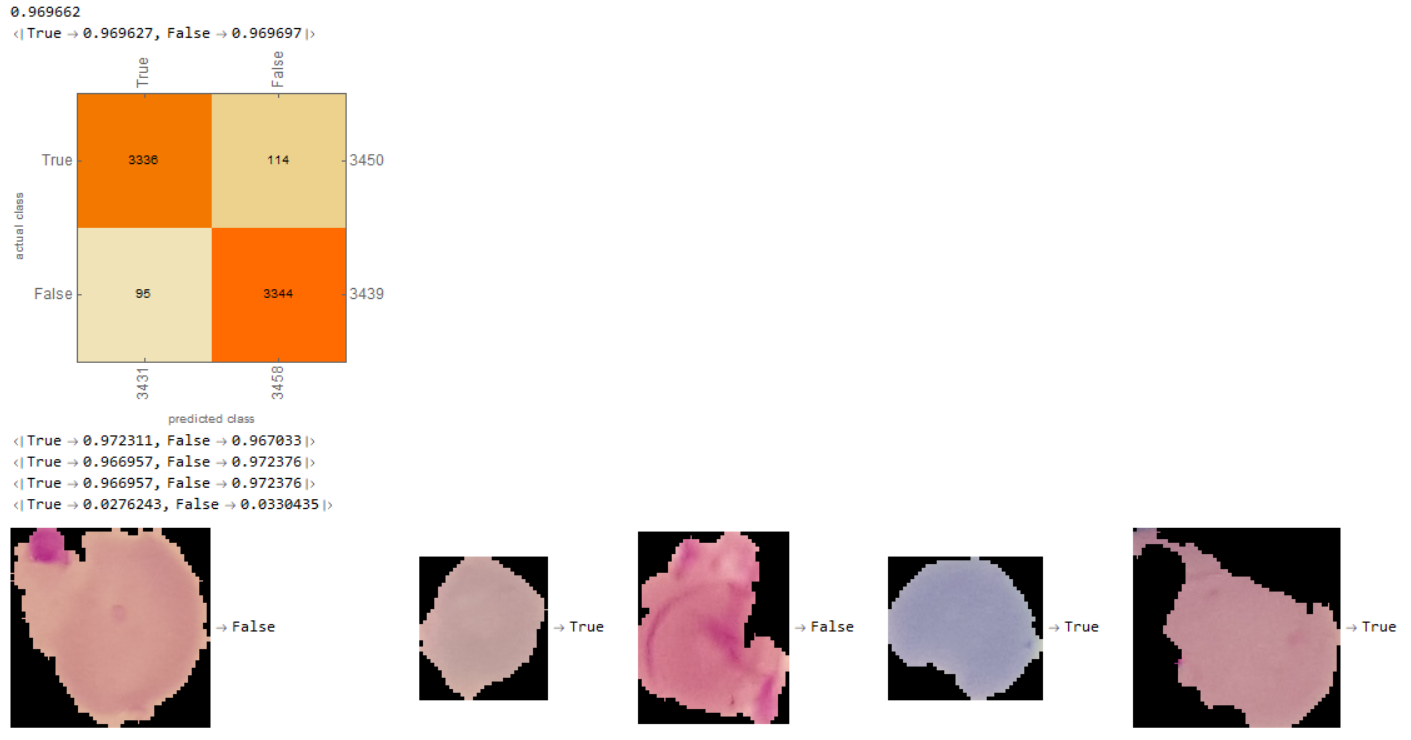
Conclusion
I created a neural network that successfully diagnosed Malaria with an accuracy of about 97%. Furthermore, as displayed in the ConfusionMatrix, there were 3336 examples of the neural network prediction matching with the actual results for true and 3344 examples of the neural network and actual matching for false. Thus, the neural network was relatively even in predicting classes and didnt favor one class over the other.
Acknowledgement
I would like to thank my mentor, Emma Yang, for helping guide me through this process. I would also like to thank other mentors including, Sylvia Haas and Mohammad Bahrami for further assistance throughout my project. Finally, I would like to thank my parents and older brother for supporting me and pushing me to keep learning.
Future Improvements
To further improve this project, I could implement more augmented data sets to further train and improve the neural net. Furthermore, I could use different cell images from different data sets to prevent over fitting and increase accuracy. Lastly, I could implement a function that pinpoints the malaria in the blood cell by finding the edges of the cell and sensing the infected cells through the function, image partition and color detection.