If you have version 12.1, then you can use the OpenCascade Link to give access to the open source CAD program. Like most CAD packages, it can do a good job with boolean operations.
I am by no means an expert, but I was able to throw this workflow together based on the tutorial.
(* Load Required Packages *)
Needs["OpenCascadeLink`"]
Needs["NDSolve`FEM`"]
(* parameters *)
z0 = 3.1981; z1 = 0.9; z2 = -0.036; z3 = 0.001;
lowz = 0.0001;
highz = 9.05;
mainR = (z0 + z*(z1 + z*(z2 + z3*z))) /. z -> highz;
highPinz = 4.88;
(* create curve for LED *)
pp = ParametricPlot[{(z0 + z*(z1 + z*(z2 + z3*z))), z}, {z, lowz,
highz}, AspectRatio -> Automatic, PlotRange -> {{0, All}, All},
MaxRecursion -> 1, PlotPoints -> 20]
(* Extract points from plot *)
coordinates = First[Cases[Normal[pp], Line[l_] :> l, \[Infinity]]];
(* Convert coordinates to 3D *)
pts = ArrayPad[coordinates, {{0, 0}, {0, 1}}];
pts[[All, {1, 2, 3}]] = pts[[All, {1, 3, 2}]];
(* Create surface of revolution in OpenCascade *)
ll = Line[pts];
wire = OpenCascadeShape[ll];
axis = {{0, 0, 0}, {0, 0, 1}};
sweep = OpenCascadeShapeRotationalSweep[wire, axis];
bmesh = OpenCascadeShapeSurfaceMeshToBoundaryMesh[sweep];
Show[Graphics3D[{{Red, Thickness[0.02], ll}, {Blue, Thick,
Arrow[10 axis]}}], bmesh["Wireframe"], Boxed -> False]
(* Cap ends of swept shell *)
crd = bmesh["Coordinates"];
lowcap = Polygon@Select[crd, (#[[3]] == lowz) &];
highcap = Polygon@Select[crd, (#[[3]] == highz) &];
p1 = OpenCascadeShape[lowcap];
p2 = OpenCascadeShape[highcap];
shape = OpenCascadeShapeSewing[{sweep, p1, p2}];
(* Create main body *)
cylshape =
OpenCascadeShape[
c1 = Cylinder[{{0, 0, lowz}, {0, 0, highPinz}}, mainR]];
(* Difference LED from main in OpenCascade *)
diff = OpenCascadeShapeDifference[cylshape, shape];
(* Visualize *)
bmeshshape = OpenCascadeShapeSurfaceMeshToBoundaryMesh[shape];
bmeshcyl = OpenCascadeShapeSurfaceMeshToBoundaryMesh[cylshape];
bmeshdiff = OpenCascadeShapeSurfaceMeshToBoundaryMesh[diff];
groups = bmeshdiff["BoundaryElementMarkerUnion"];
temp = Most[Range[0, 1, 1/(Length[groups])]];
colors = ColorData["BrightBands"][#] & /@ temp;
Show[bmeshshape["Wireframe"], bmeshcyl["Wireframe"]]
bmeshdiff["Wireframe"["MeshElementStyle" -> FaceForm /@ colors]]
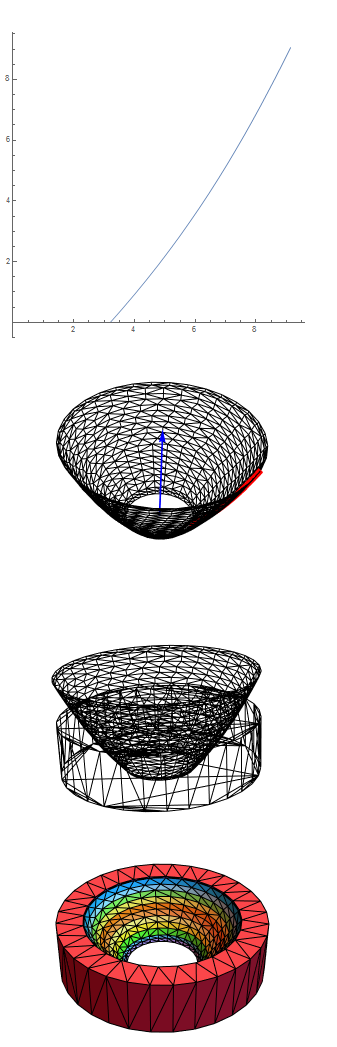
It appears that OpenCascade does a good job maintaining sharp edges.