I am a neophyte to WL and MM (and honestly this is my first programming language) and I hope to learn, from WL/MM experts or advanced peers, how to programmatically think about the following image-processing tasks:
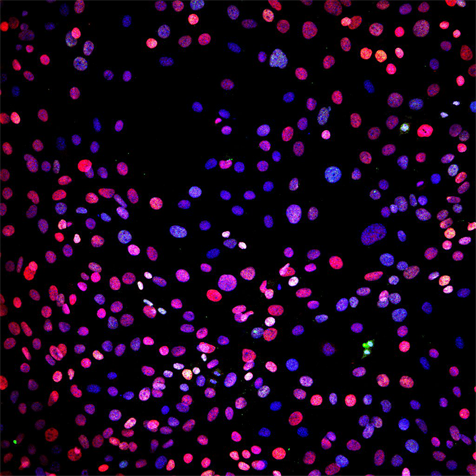
A) to calculate the higher-order statistical moments (SD, skewness, and kurtosis) for the intranuclear (blue) distribution of protein1 (red) and protein2 (green). Image entropy can be calculated as E(X) = ?i?1p(x)log(p(x)) over all of the possible states of x of ?, where p(x) is the probability of state x occurring within the image. Entropy can be calculated by the selection of 500 random pixels (states) within each nuclei to avoid bias resulting from changes in the number of selected pixels and pixel with ?-intensities rounded to the nearest 0.001.
B) to perform pixel intensity spatial correlation analysis using Pearson's correlation coefficient or Manders split coefficients or alternatively using the Euclidean distance.
PS: Although I am only 6 months old in WL/MM/programming I tried to write some code to address (B) but I did not get any reply or comments .Please see:
http://community.wolfram.com/groups/-/m/t/956956
That's actually one self-contained problem: How to spatially colocalize two signals?
I went through many tutorials and documentation and even drove 11 hours back and forth to attend the the 2015 image-processing workshop in Urbana-Champaign (with my modest graduate student means). I spend several weeks trying to figure out this without asking anyone helps and emailed your technical service many times but with no consideration or answers.
Please kindly provide any link of a tutorial or an image-processing documentation link that address :
- Intensity Spatial Correlation otherwise known as co-localization in fluorescence microscopy.
- Correlation based on any image distance.
I am experimenting in vitro with stem cells because they transitions between different stages in a cellular automata -like fashion. the same rationale tried in vivo seems to be consistent with my in vitro model and with many of the NKS ideas. PS: I will move on to human cells in vitro soon.
Regarding the distance. I am talking image distance like the Euclidean are Manhattan distance see
http://community.wolfram.com/groups/-/m/t/958365
Many thanks in advance for any guidance I can get for the Wolfram community.