A new SystemModeler example has been added to the list of downloadable models available on the SystemModeler site, Enzyme Catalysis.
Enzymes play a part in almost every biochemical reaction known, from human metabolism to beer brewing. This model showcases a generic substrate to product reaction and catalyzes it to illustrate the power of enzymes. The free BioChem Modelica library is needed to run this example; it can be downloaded here.
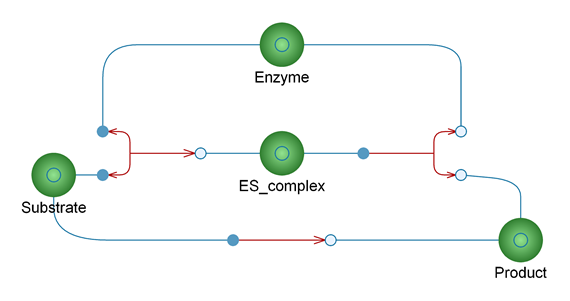
In the example, dynamic interactivity using the Wolfram Language is explored. Interactive objects, such as sliders can be really helpful in trying to understand the concept that the model explores. This has been made all the more possible with the performance increases in the SystemModeler 4.2 and 4.3 releases. In this example, you can use sliders to explore how enzyme concentration and substrate concentration affects the product formation in the system.
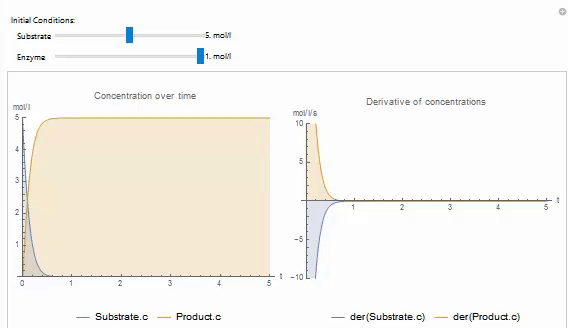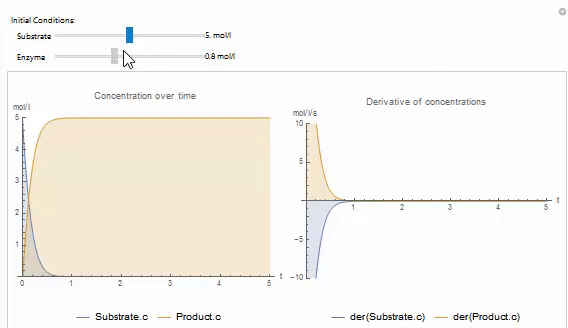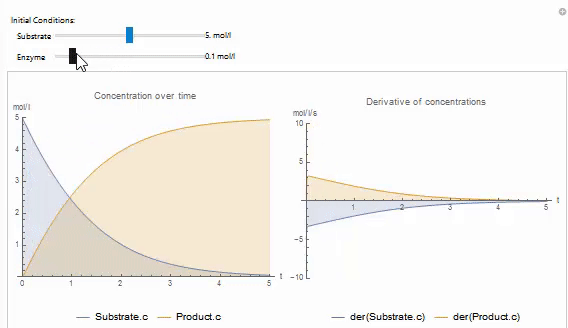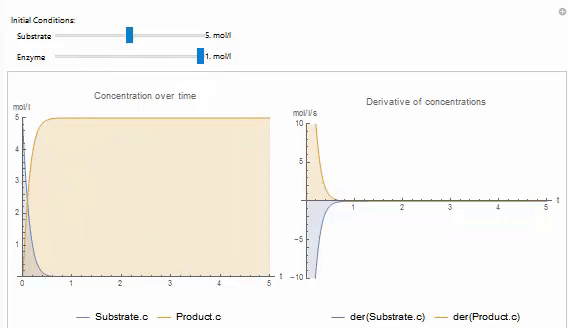
You can easily create your own Wolfram Language control objects to explore your models. For example, the following code produces a manipulate that can be used to set the initial value of a variable in on of the built in examples:
Needs["WSMLink`"]
Manipulate[WSMPlot[
WSMSimulate["HelloWorld", WSMInitialValues -> {"x" -> x0}]
],
{x0, 0, 10},
ContinuousAction -> False]
It is recommended to set ContinuousAction to False so the simulation will only be run when the sliders are dropped, not continuously while dragging the slider.