Great work and very usefull!
I have generalized your network such that it can take any number of channels and classes. Your case handles one channel and one class. However i did not manage to make the single class output a binary image since the SoftmaxLayer and NetDecoder do not allow to classify one class and i did not find a suitable solution. The SoftmaxLayer with class labeling allows to give error rate feedback during the training.
pool := PoolingLayer[{2, 2}, 2];
upSamp[n_] := DeconvolutionLayer[n, {2, 2}, "Stride" -> {2, 2}]
conv[n_, p_: 0] := NetChain[{If[p == 1, pool, Nothing],
ConvolutionLayer[n, 3, "PaddingSize" -> {1, 1}],
BatchNormalizationLayer[], Ramp,
ConvolutionLayer[n, 3, "PaddingSize" -> {1, 1}],
BatchNormalizationLayer[], Ramp
}];
dec[n_] := NetGraph[{
"deconv" -> DeconvolutionLayer[n, {2, 2}, "Stride" -> {2, 2}],
"cat" -> CatenateLayer[],
"conv" -> conv[n]},
{NetPort["Input1"] -> "cat",
NetPort["Input2"] -> "deconv" -> "cat" -> "conv"}
];
UNET2D[NChan_: 1, Nclass_: 1] := NetGraph[<|
"enc_1" -> conv[64], "enc_2" -> conv[128, 1],
"enc_3" -> conv[256, 1], "enc_4" -> conv[512, 1],
"enc_5" -> conv[1024, 1],
"dec_1" -> dec[512], "dec_2" -> dec[256], "dec_3" -> dec[128],
"dec_4" -> dec[64],
"map" -> {ConvolutionLayer[Nclass, {1, 1}], LogisticSigmoid,
If[Nclass > 1, TransposeLayer[{1 <-> 3, 1 <-> 2}], Nothing],
If[Nclass > 1, SoftmaxLayer[], Nothing],
If[Nclass > 1, Nothing, FlattenLayer[1]]}
|>,
{NetPort["Input"] ->
"enc_1" -> "enc_2" -> "enc_3" -> "enc_4" -> "enc_5",
{"enc_4", "enc_5"} -> "dec_1", {"enc_3", "dec_1"} -> "dec_2",
{"enc_2", "dec_2"} -> "dec_3", {"enc_1", "dec_3"} -> "dec_4",
"dec_4" -> "map"},
"Input" -> {NChan, 128, 128},
"Output" ->
If[Nclass > 1,
NetDecoder[{"Class", "Labels" -> Range[1, Nclass],
"InputDepth" -> 3}], Automatic]
]
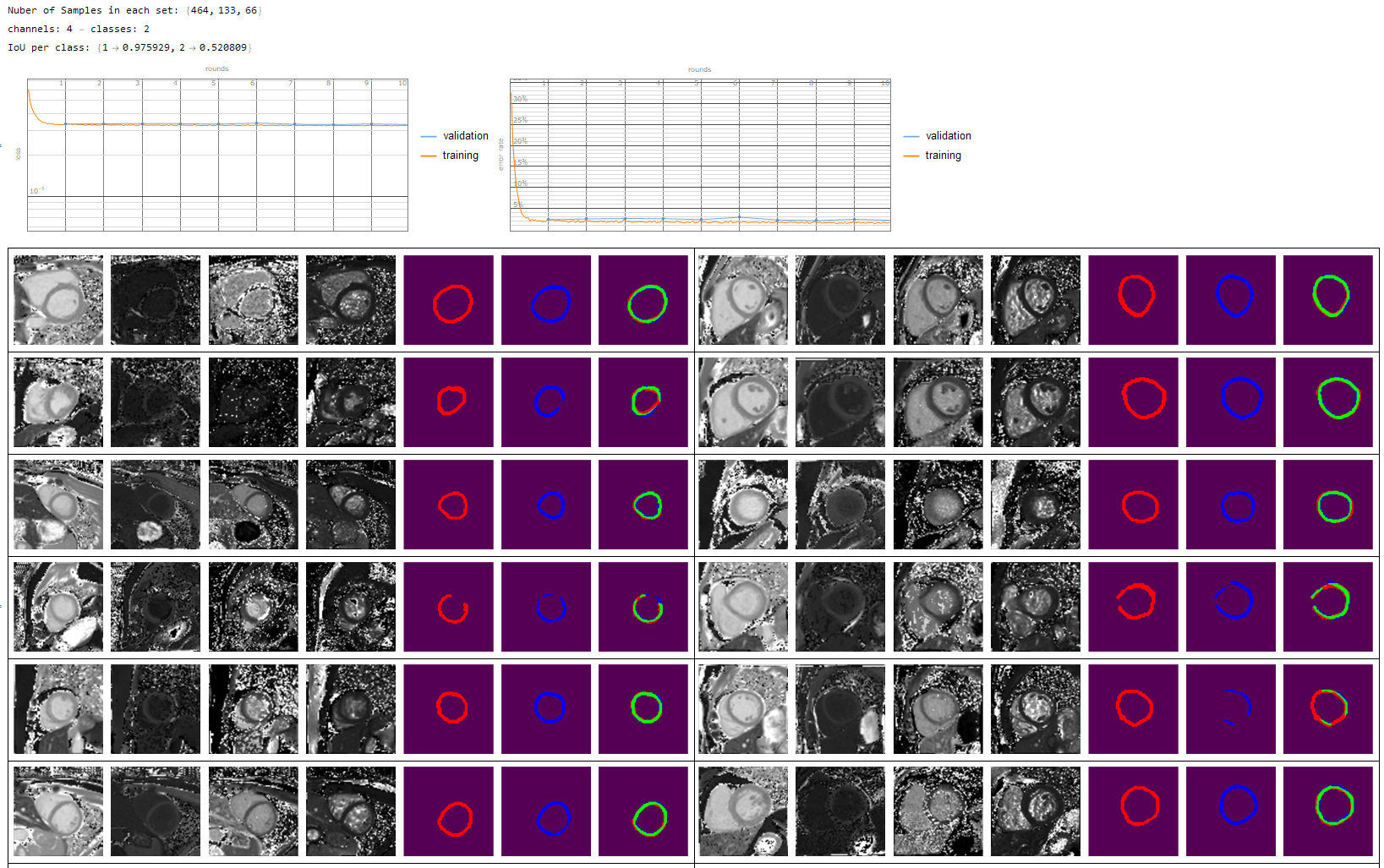
I also made the IuO function a bit faster for large data-sets and it reports the values per calss
ClassIOU[predi_, gti_, class_] :=
Block[{posN, posP, fn, tp, fp, denom},
(*find posisions as unitvector*)
posN = Unitize[predi - class];
posP = 1 - posN;
(*get the values*)
fn = Count[Pick[gti, posN, 1], class];
tp = Count[Pick[gti, posP, 1], class];
fp = Total[posP] - tp;
(*if case does not exist in all data return 1*)
denom = (tp + fp + fn);
If[denom == 0, 1., N[tp/denom]]
]
IOU[pred_, gt_, nClasses_] := Block[{predf, gtf},
predf = Flatten[pred];
gtf = Flatten[gt];
Table[ClassIOU[predf, gtf, c], {c, nClasses}]
]
To split the available data into training, validation and testing data I made this function.
SplitTestData[data_, label_, ratio_: {0.7, .2, .1}] :=
Block[{allData, train, valid, test, testData, testLabel, s1, s2, s3},
(*Randomize data*)
allData = RandomSample[Thread[data -> label]];
(*split data*)
Print["Nuber of Samples in each set: ", Round[ratio Length[allData]]];
{s1, s2, s3} = Accumulate@Round[ratio Length[allData]];
(*make training validation and test data*)
train = allData[[1 ;; s1]];
valid = allData[[s1 + 1 ;; s2]];
test = allData[[s2 + 1 ;;]];
testData = test[[All, 1]];
testLabel = test[[All, 2]];
(*define the output*)
{train, valid, testData, testLabel}
]
These are some generated test dataset on which i tested the network
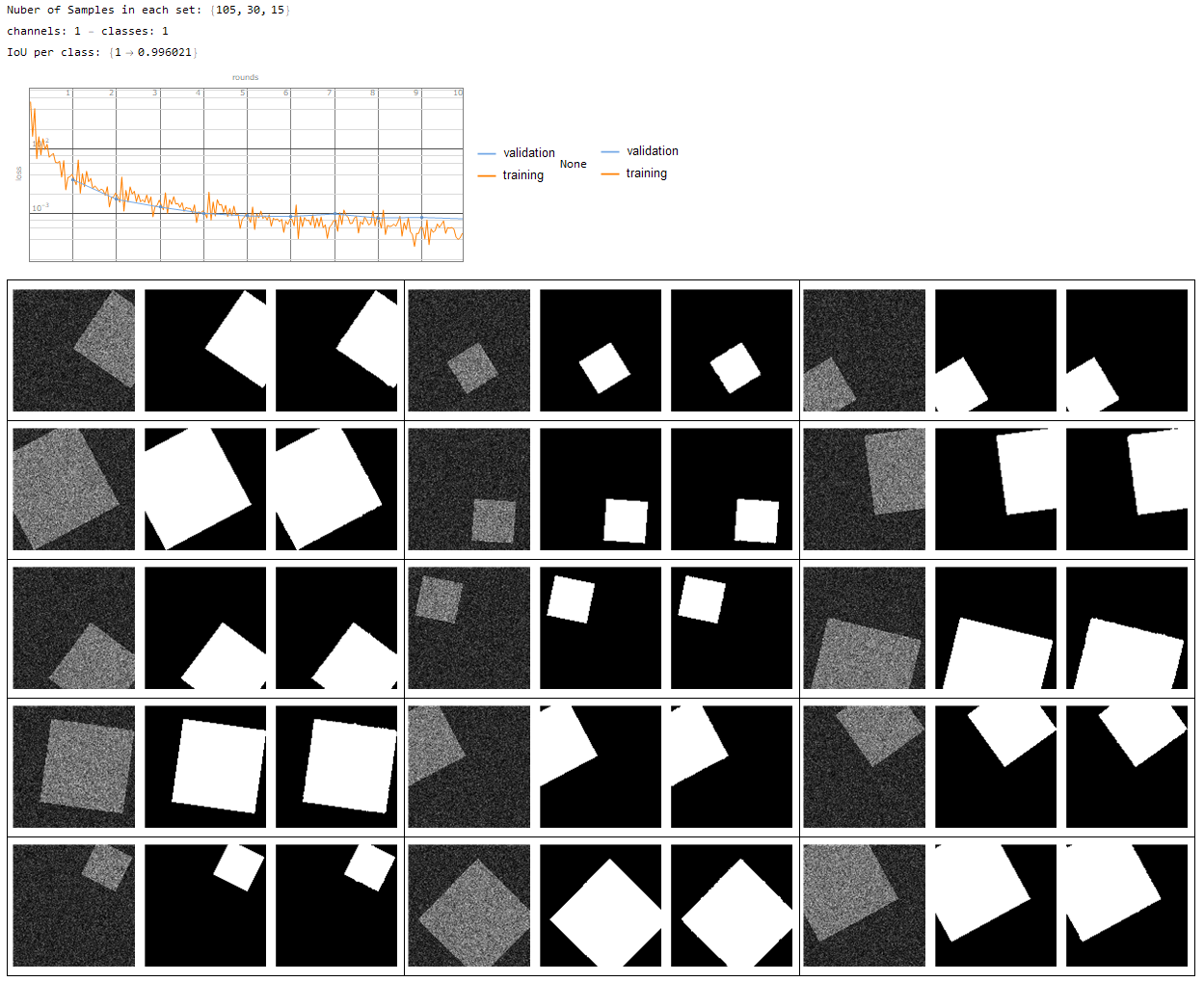
- single channel - single class
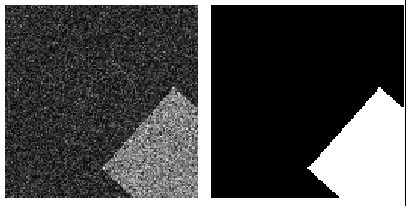
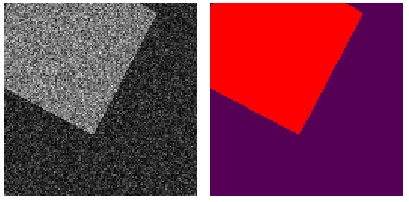
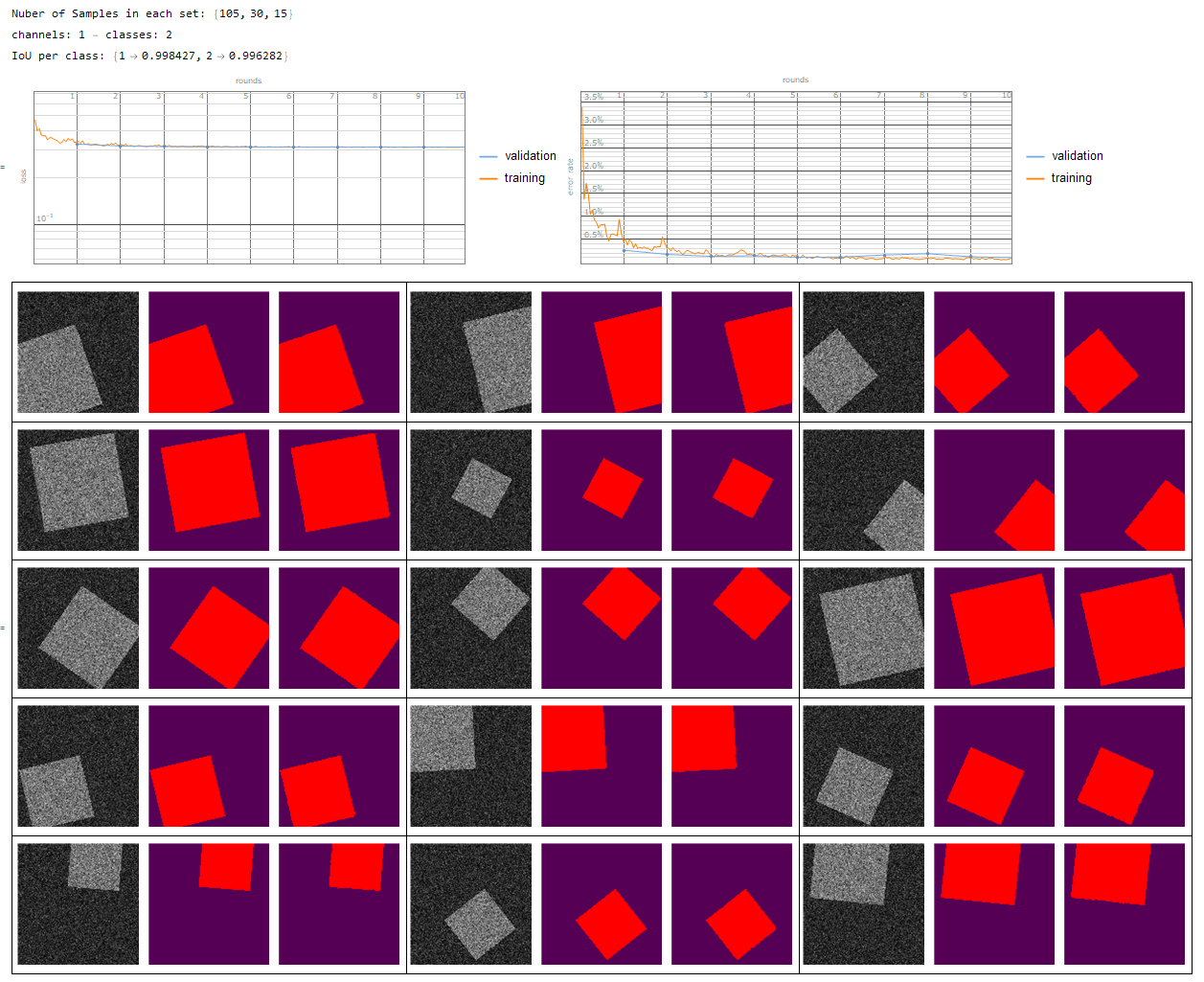
- single channel - multi class
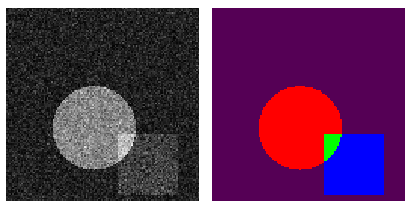
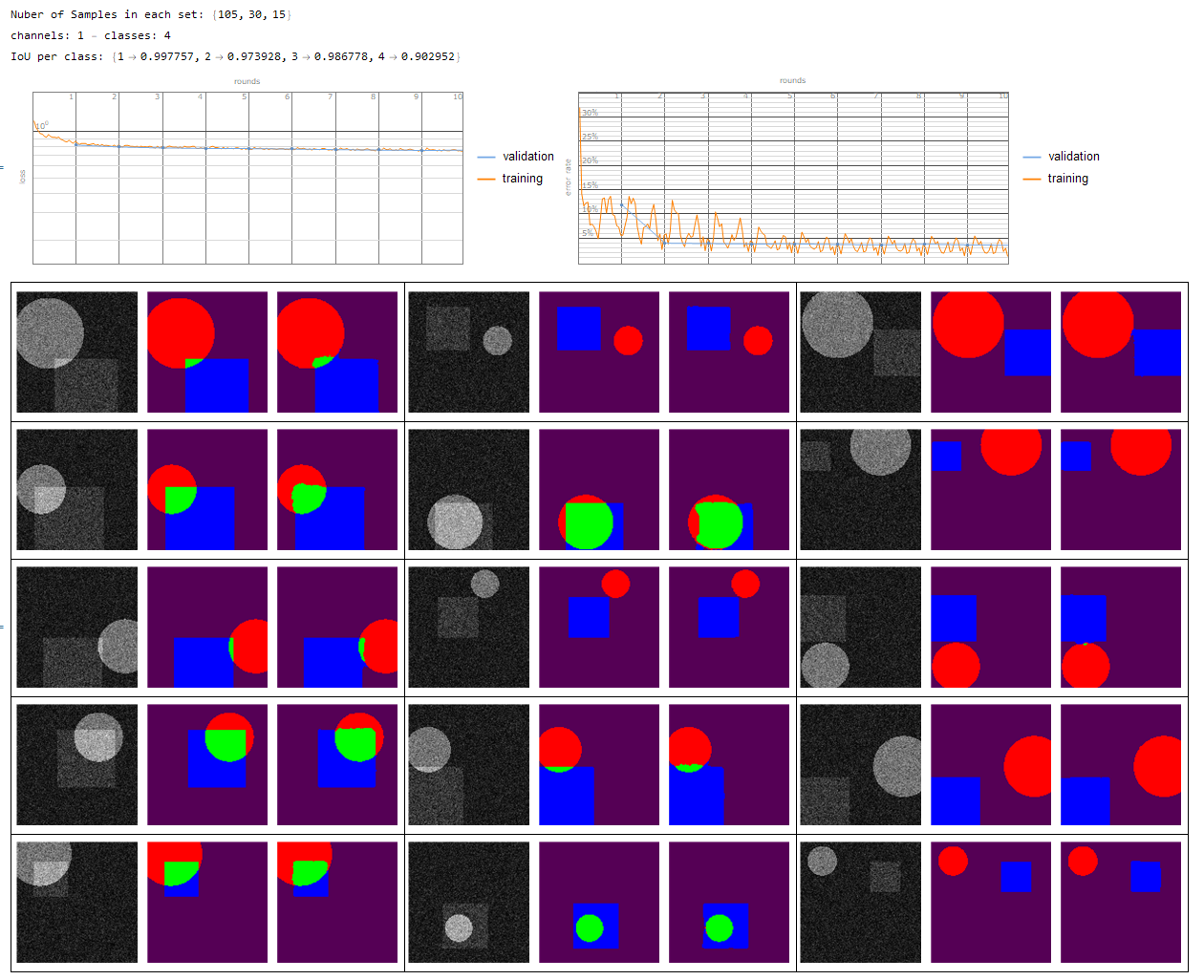
- multi channel - multi class
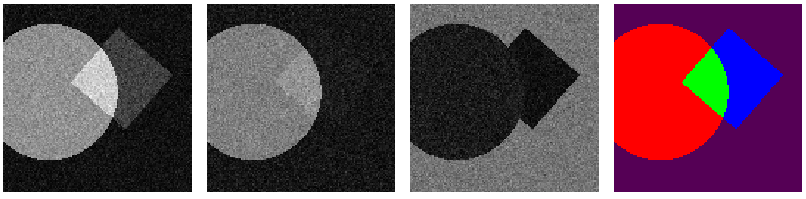
These were the results
- single channel - single class
- single channel - 2 classes {1 -> Background, 2 -> segmentation}
- single channel - multi class {1 -> Background, 2...n -> segmentation}
- multi channel - multi class {1 -> Background, 2...n -> segmentation}
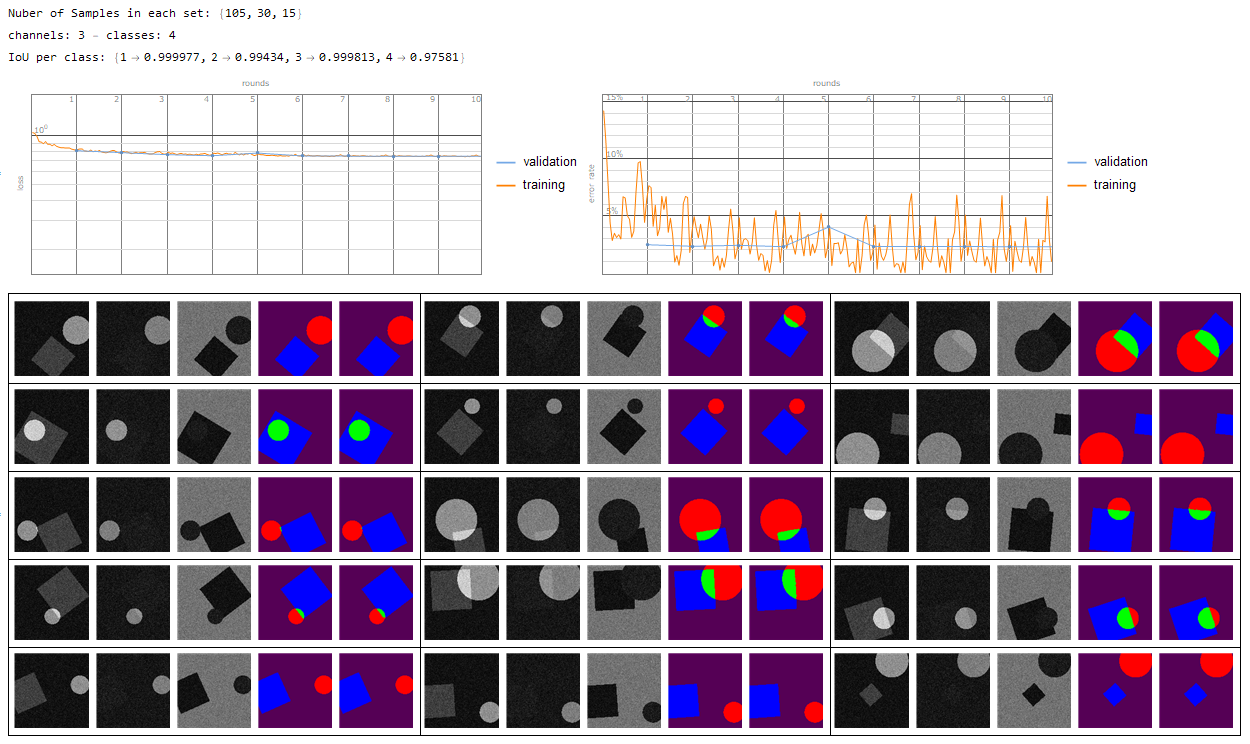
And finally of course my real application to segment the heart wall from multi modal MRI contrasts. However id do have to admit that my manual annotations could be a bit better such that the network can train more accurately.
(Red - Manual annotation, Blue - Trained net, Green - overlap of the two)
 Attachments:
Attachments: