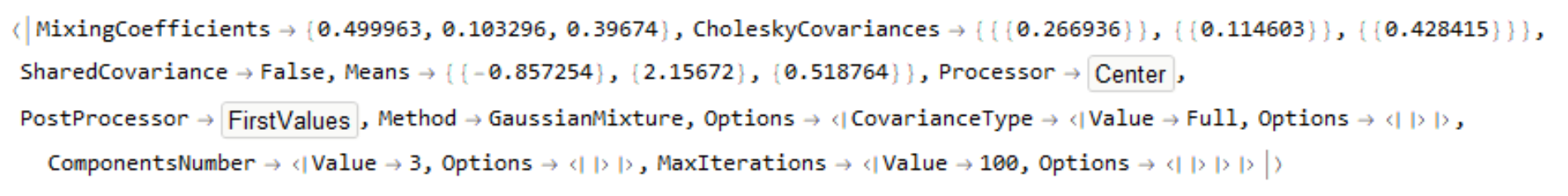
Not because LearnDistribution is Experimental but because there aren't many details about what it actually does is why I think you should shy away from it. One can "extract" the parameters used but I don't think that all of those parameters make sense. For example one of the means is negative when you have all positive data with the leftmost of 3 "peaks" greater than zero.
data = {0.563000, 0.180000, 0.429000, 0.292000, 0.315000, 0.888000,
0.59100, 0.196000, 0.200000, 0.581000, 0.265000, 0.288000,
0.492000, 0.600000, 0.722000, 0.471000, 0.232000, 0.835000,
0.370000, 0.265000, 0.266000, 0.340000, 0.239000, 0.931000,
0.201800, 0.522000, 0.635000, 0.537000, 0.503000, 0.201000,
0.438000, 0.627000, 0.356000, 0.462000, 0.156000, 0.875000};
(* LearnDistribution *)
ld = LearnDistribution[data, Method -> {"GaussianMixture", "ComponentsNumber" -> 3,
"CovarianceType" -> "Full"}]
ld[[1, 6]]
I suggest fitting a mixture of Gaussian distributions in a more direct way:
(* Maximum likelihood approach *)
md = MixtureDistribution[{w1, w2, 1 - w1 - w2}, {NormalDistribution[m1, s1],
NormalDistribution[m2, s2], NormalDistribution[m3, s3]}]
mle = FindDistributionParameters[data, md, ParameterEstimator -> "MaximumLikelihood"]
(* {w1 -> 0.467002, w2 -> 0.110224, m1 -> 0.524247, s1 -> 0.0988225,
m2 -> 0.882558, s2 -> 0.0341755, m3 -> 0.246329, s3 -> 0.0562667} *)
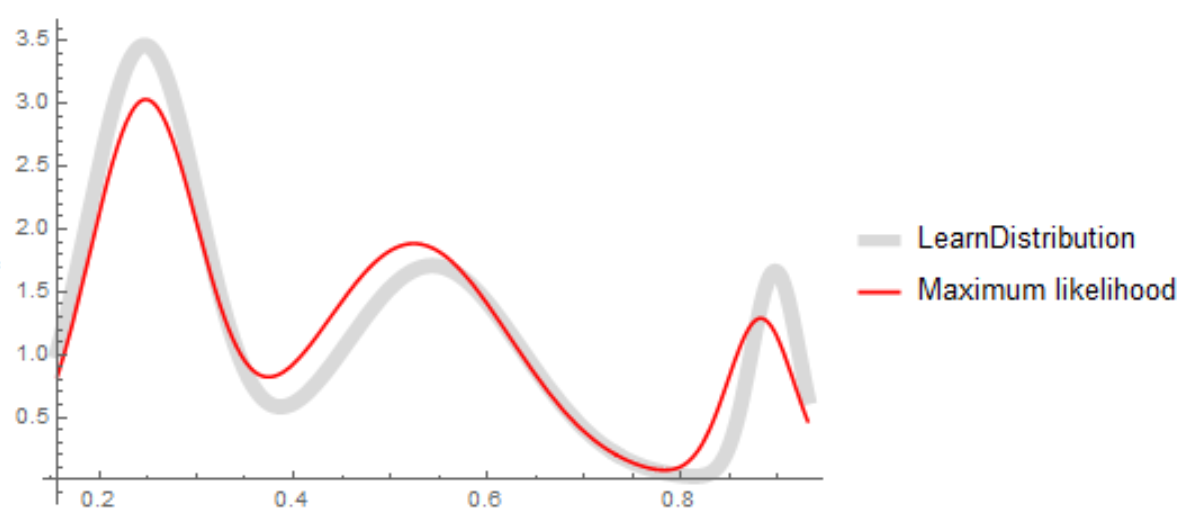
The results are not identical but at least you know how you got the maximum likelihood results:
(* Plot of results *)
Plot[{PDF[ld, x], PDF[md, x] /. mle}, {x, Min[data], Max[data]},
PlotStyle -> {{LightGray, Thickness[0.02]}, Red},
PlotLegends -> {"LearnDistribution", "Maximum likelihood"}]
Your original question is to get the estimate of the PDF. Here's that function:
PDF[md/.mle]
Function[\[FormalX],
1.28668 E^(-428.094 (-0.882558 + \[FormalX])^2) +
1.88527 E^(-51.1986 (-0.524247 + \[FormalX])^2) +
2.99756 E^(-157.931 (-0.246329 + \[FormalX])^2)]
Now back to your original question: How to get the parameters associated with the results from LearnDistribution? Again, I think you should avoid using LearnDistribution not because it's bad but because it's a black box with inadequate documentation. However, one can get the associated parameters at least in a reasonably precise way. We can generate a table of a bunch of values and use NonlinearModelFit because we know the variability about the curve is going to be very small.
t = Table[{x, PDF[ld, x]}, {x, Min[data], Max[data], (Max[data] - Min[data])/100}];
nlm = NonlinearModelFit[t, PDF[md, x], mle /. Rule -> List, x];
fit = nlm["BestFitParameters"]
(* {w1 -> 0.39674, w2 -> 0.103296, m1 -> 0.543333, s1 -> 0.0926299,
m2 -> 0.897484, s2 -> 0.0247789, m3 -> 0.245816, s3 -> 0.0577158} *)
Plot[{PDF[ld, x], PDF[md, x] /. mle, PDF[md, x] /. fit}, {x, Min[data], Max[data]},
PlotStyle -> {{LightGray, Thickness[0.02]}, Red, Green},
PlotLegends -> {"LearnDistribution", "Maximum likelihood", "NonlinearModelFit"}]

So, finally, the pdf is
PDF[md,x]/.fit
1.66308 E^(-814.34 (-0.897484 + x)^2) +
1.7087 E^(-58.273 (-0.543333 + x)^2) +
3.45584 E^(-150.1 (-0.245816 + x)^2)