Version 12 of the Wolfram Language introduced the Molecule object type, which represents a molecule as a chemical graph with atoms, bond, stereochemistry, with specified 2D and 3D coordinates. The MoleculeModify function lets you programmatically alter the geometry via the "SetBondLength", "SetBondAngle", and "SetTorsionAngle" methods.
Now there is a function for modifying a molecule's 3D coordinates interactively: MoleculeManipulate3D - a part of the ChemistryFunctions paclet available from the Wolfram Language Paclet Repository.
To install the paclet use
PacletInstall["WolframChemistry/ChemistryFunctions"]
To load the paclet in a kernel session use
Needs["WolframChemistry`ChemistryFunctions`"]
Doing this will make all of the functions in the paclet available. Then use something simple like
m = Molecule["octane"];
MoleculeManipulate3D[m]
and you will see this dynamic output:
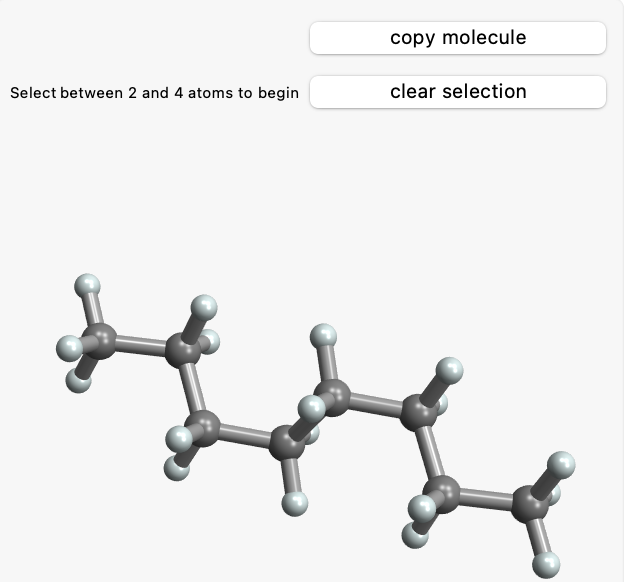
- Once a first atom is selected, click that atom again to deselect it or click any atom that is bonded to the first atom to select two atoms. Clicking anywhere else will not change the selection.
- When two bonded atoms are selected a slider will appear that will change the given bond length. If the bond is part of a ring system the slider will not be enabled.
- When three atoms are selected a slider will appear which allows changing the given bond angle. If both bonds are part of a ring system the slider will not be enabled.
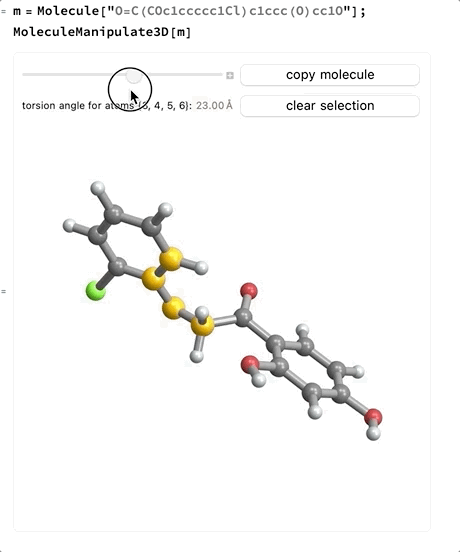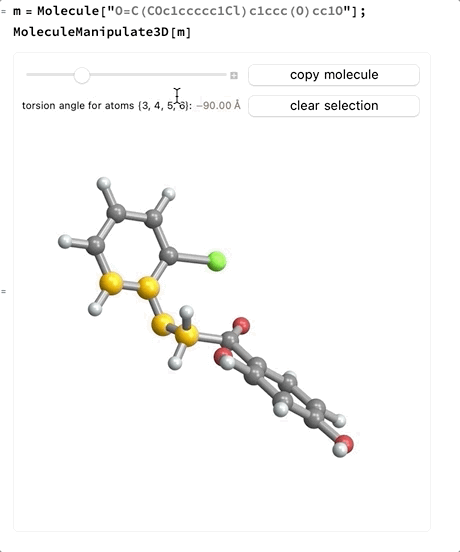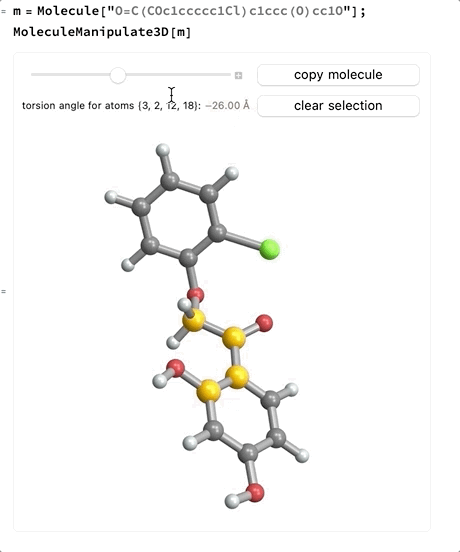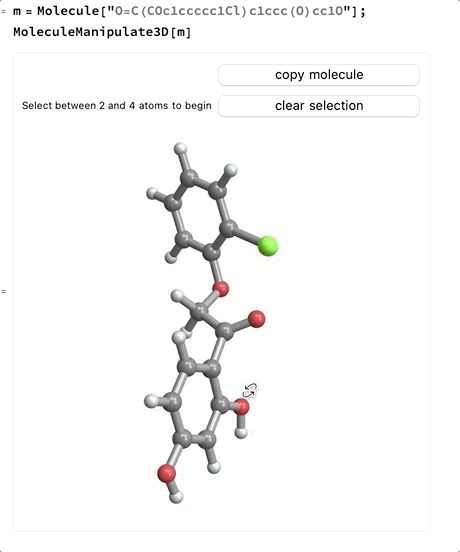
- When four atoms are selected a slider will appear which allows changing the signed torsion angle between the plane made by the first three atoms and the plane made by the last three atoms. If the bond between the second and third atoms is part of a ring system the slider will not be enabled.
Unfortunately the dynamics used in this function do not currently work in Wolfram Cloud notebooks.