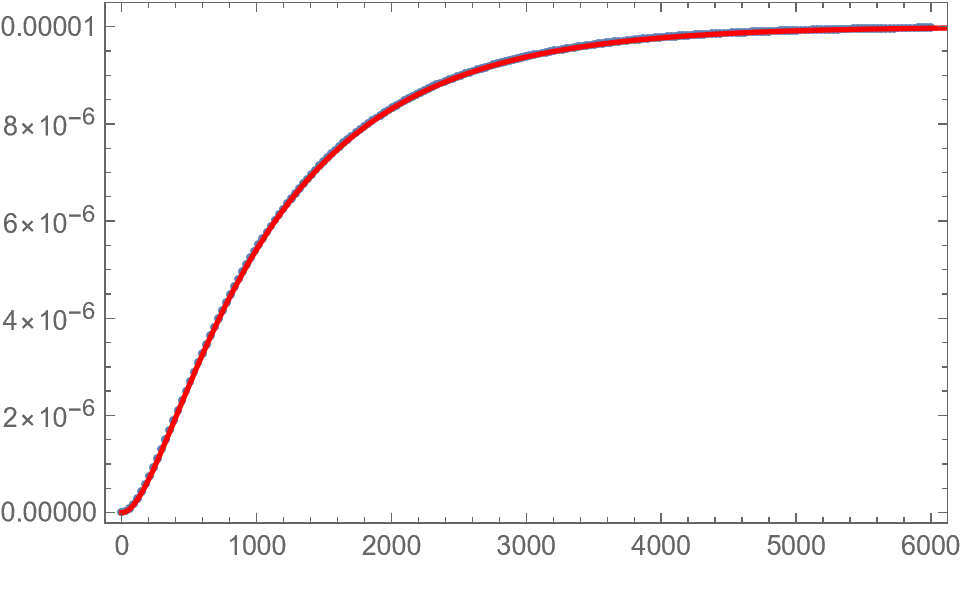
That message is a "warning" rather than an "error" as you see you still get a good fit. However, the underlying reason for the warning is that your model is overparametersized. If you look at the parameter correlation matrix you see the following:
nlf["CorrelationMatrix"]

The correlations for parameters Kt, K3, and Kmeno3 are all either +1 or -1 indicating that the model is overparameterized. Essentially there is just a single parameter that can be estimated among those 3 parameters:
model //. K3 Kt -> a (Kmeno3 + Kt) // FullSimplify
(* (c (a - a E^(-Kp x) + (-1 + E^(-a x)) Kp))/(a - Kp) *)
This more parsimonious model gives the exact same predictions as your original model but (likely) with fewer numerical issues. The only issue is that starting values can't be the same for a and Kp otherwise the denominator will start off as zero.
nlf2 = NonlinearModelFit[data, (c (a - a E^(-Kp x) + (-1 + E^(-a x)) Kp))/(a - Kp),
{a, c, {Kp, 0.0001}}, x];
nlf2["BestFitParameters"]
(* {a -> 0.00500545, c -> 0.0000100002, Kp -> 0.000999561} *)
Show[ListPlot[data],
Plot[nlf[x], {x, 0, end}, PlotStyle -> Red, PlotRange -> All],
Frame -> True]