skinks = {56, 19, 28, 18, 24, 14, 9}; kskinks = 7;
myBBBlik[data_, kdata_, f0_?NumericQ, p_?NumericQ, w_?NumericQ, alpha_?NumericQ, beta_?NumericQ] :=
Block[
{K, fj, pj, loglik},
K = kdata;
fj = Prepend[PadRight[data, K], f0];
pj = Table[
PDF[MixtureDistribution[{w, 1 - w}, {BinomialDistribution[K, p],
BetaBinomialDistribution[alpha, beta, K]}], j],
{j, 0, K}
];
loglik = LogGamma[Tr[fj] + 1] - Tr[LogGamma[fj + 1]] + Tr[fj*Log[pj]];
Print[loglik];
loglik
];
pars = {f0,p,w,allpha,beta};
cons = {f0 > 0 && 0 < p < 1 && 0 < w < 1 && alpha > 0 && beta > 0};
cons2 = {f0 > 0 , 0 < p < 1 , 0 < w < 1 , alpha > 0 , beta > 0};
NMaximize[{myBBBlik[skinks, kskinks, Sequence @@ pars], cons}, pars, Method -> {"RandomSearch", "SearchPoints" -> Automatic}]
Note that it may crash your kernel!
If you take out the line Print[loglik];, it will give your the correct answer. Works perfect. However, if you leave it in and it may crash your kernel, but you will see something (unbelievable && unexpected) like this: 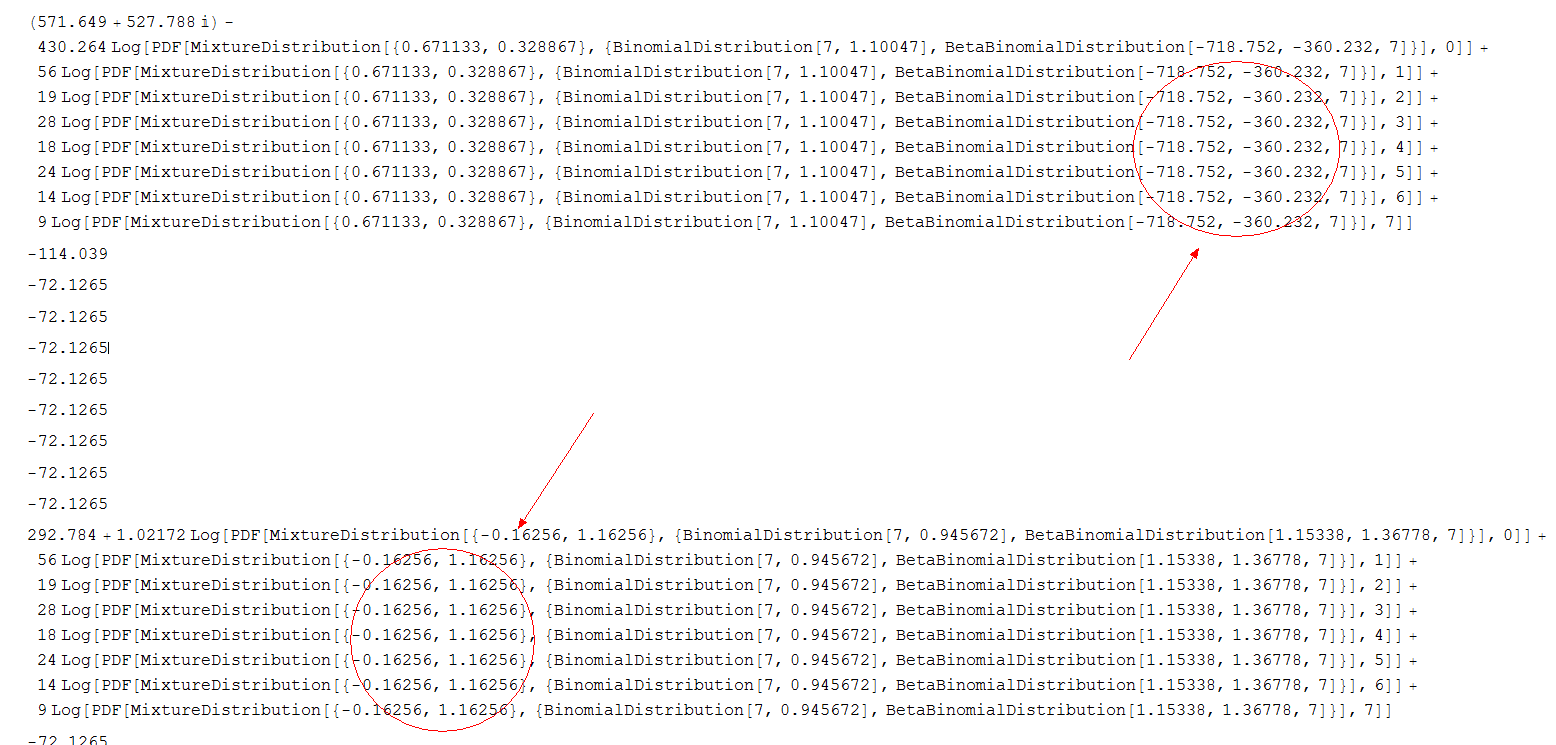
Why does it even call the function myBBBlik? Values like -717.752 and -360.232 are long way from 0. These would not considered when you have alpha>0,beta>0. This is just really something I don't understand the way MMA works.
I am using MMA 10.0.2 X64 (win)