I'm interested in motion capture data. We understand musculature, yet motion capture remains a tool used only for skeletal animation. I suspect proper interpretation of mocap data could create tools for diagnosis. To extrapolate muscle interaction from skeletal data requires knowledge of how muscles connect. This mid level data seems elusive compared to both high level understanding, and low level raw scans.
I found model data for each muscle in the body. My goal is to compress this dense data on the assumption that all muscles are essentially splines joining their end points.
The plan is to use surface traversals of the model vertices to calculate the rough shape of the muscle. Then to adjust this shape back towards the vertex cloud to remove inaccuracies introduced by the first step.
This is the raw data of a muscle I'm starting with. First it's polygonal render, then the vertices that make up the render.

I use two functions to convert the point cloud into a single progression. The first finds the shortest path through a set of points, and partitions that path into pairs; the pairs are then averaged to form a smaller point set that closely resembles the first. The second function takes a degree five spline through a progression of points, and returns the points projected onto the spline. This is a low cost way to smooth a discrete point progression. Corners in the progression of higher angle are adjusted further than points in linear progression.
By repeatedly applying both of these procedures, the surface point cloud is rapidly reduced to a short progression with low curvature.
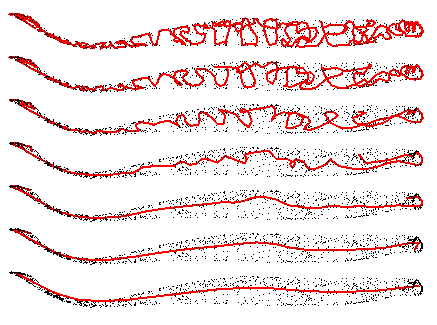
The spline smoothing inherently cuts corners. This is visible above, by the seventh revision the approximation has left the point cloud in the thin, curved section. Whilst spatially inaccurate, the approximation does tell us what direction the muscle progresses in. Armed with this information, the muscle can be dissected along its length, and compressed into a set of 2D slices. This is done by finding the point on the spline closest to each vertex, which in turn orders the vertices along the progression of the muscle.

The dimensional extrema of these 2D point sets give good oblong approximations along the length of the muscle, a data reduction of about 20 fold over the original point cloud. Averaging the extrema gives the final progression. A very simple and accurate parameterisation of the muscle!
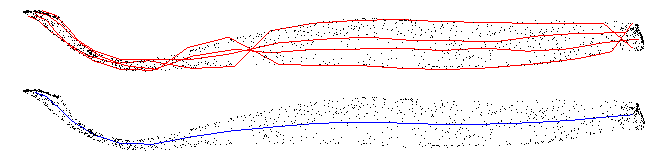
This procedure is far from perfect. I've spent a long time bashing bugs and failures. A large component is the time taken to find the closest point on the spline from a vertex. I'm using a high degree spline with a single equation, and evaluating distance at all inflection points. Whilst this is accurate, I wonder if anyone could suggest faster approximations.
Thanks for reading!
David
 Attachments:
Attachments: