Hi all, as an attempt to model a process in molecular biology, I have put together a set of differential equations to describe changes in the concentration of certain proteins (PhaC), as they go through an initial complex formation (PhaCC), and multiple subsequent reactions adding one S group each time. Each S carrying complex is also subject to hydrolysis, which yields the protein complex and the number of S groups carried, as free monomers. The image below is representative.
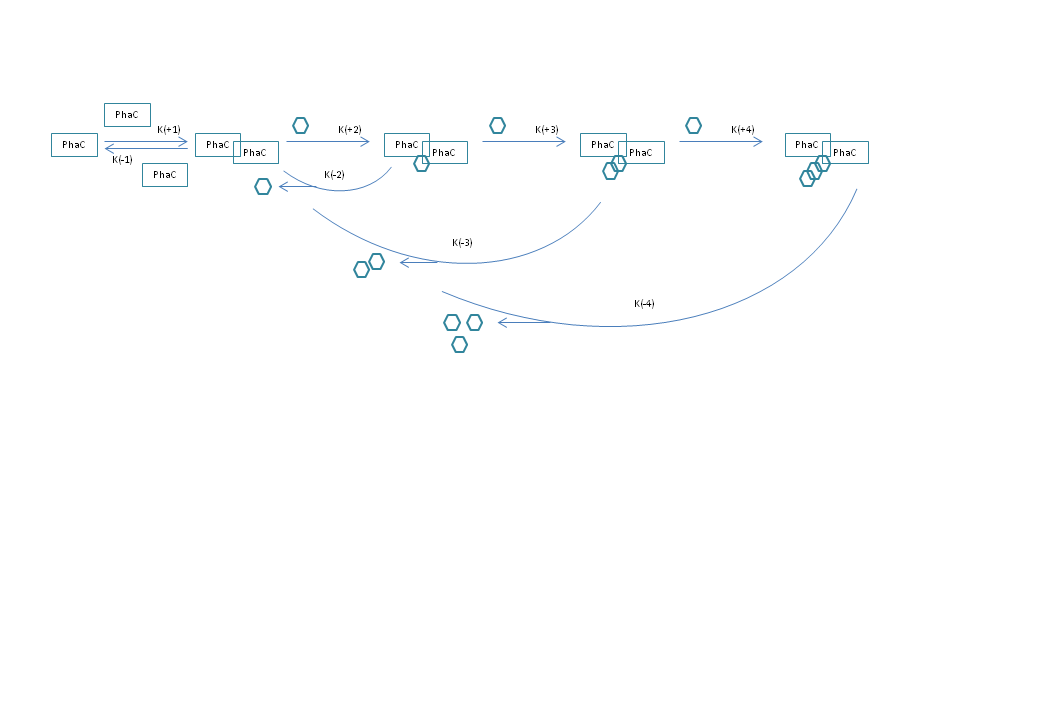
Except for the complex formation, all reactions follow irreversible mass action kinetics. Furthermore, all initial concentrations are set to 0, except for the protein monomers, and the S groups, also as monomers. I have encountered two problems:
- first, some protein concentrations take negative values from a certain point in time, and the substrate concentration, which is supposed to diminish, actually increases...
- second, I was unable to explicitely state a mass conservation equation in the system.
I've made lists containing the odes, the rate expressions (rateqs), the parameter (parm), the initial values (init) and the variables (vars); in the end, all are joined into an NDSolve expression goes as follows
tsol = NDSolve[Join[odes /. rateqs /. parm, init], vars, {t, 0, 10}]
I have attached the mathematica file. I'd greatly appreciate any thoughts
Thanks!
 Attachments:
Attachments: