Suppose one has a fairly long segment of nucleotide letters ATGC. A way to form an image, based on the Chaos Game Representation of Jeffrey, is as below. I use a pixellation level of 256x256 (2^8 x 2^8).
chars = {"A", "T", "G", "C"};
dim = 8;
makePositionsC = Compile[{{shifts, _Integer, 2}, {k, _Integer}},
Module[{posns},
posns = FoldList[Mod[2*#1 + #2, 2^k] &, Reverse@shifts];
Most[Reverse[Map[{2^k, 1} + {-1, 1}*Reverse[#] &, posns]]]
], RuntimeOptions -> "Speed", CompilationTarget -> "C"];
replace =
Dispatch[Thread[chars -> {{0, 0}, {0, 1}, {1, 1}, {1, 0}}]];
FCGR[chars_, alphabet_List, k_] := Module[
{shifts, posns, newposns},
shifts = chars /. replace;
newposns = Round[makePositionsC[shifts, k]];
Normal[SparseArray[Apply[Rule, Tally[newposns], {1}], {2^k, 2^k}]]
]
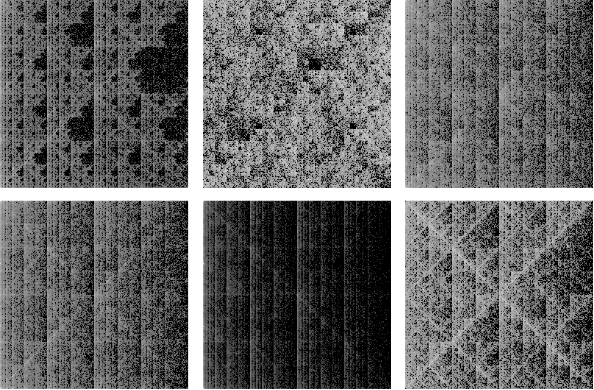
To give some idea of what gets produced, here are pictures from the six genomes discussed in one of the articles I referenced in my earlier reply. If the images are in an array images then further processing was done to get a reasonable gray scale:
scaledimages=Map[(#/N[Max[#]])^(1/6) &, images];
References
Jeffrey, H.: Chaos Game Representation of gene structure. Nucleic Acids Research 18(8), 21632170 (1990)
Lila Kari, Kathleen A. Hill, Abu S. Sayem, Rallis Karamichalis, Nathaniel Bryans, Katelyn Davis, Nikesh S. Dattani. Mapping the Space of Genomic Signatures. http://arxiv.org/abs/1406.4105
Rallis Karamichalis , Lila Kari , Stavros Konstantinidis and Steffen Kopecki, An investigation into inter- and intragenomic variations of graphic genomic signatures https://wolfr.am/fycQFg15
Wolfram Language code by the first listed coauthor can be found at the link below.
I use this in work that will be shown at SYNASC 2016 (Numerical Computing session 1). http://synasc.ro/2016/list-of-papers
Full details for obtaining the genome nucleotide sequences are in the code appendix (not sure when this will appear though), or see Rallis Karamichalis' web page.