I am reposting this here from the
Stackexchange Mathematica blog so that more people might see it. I'd be very happy to get some feedback on this plotting function. If anyone can use the function, let me know how it works out for you, and if you'd recommend any changes. If so, I can edit this post to have to most up-to-date version.
As a chemist it is often useful to plot electronic orbitals. These are used to describe the wave function of electrons in atoms or molecules. Typically, these are output from electronic structure software in the form of a cube file, first developed by Gaussian. These files contain volumetric data for a given orbital on a three-dimensional grid.
There exist many applications to visualize cube files, such as
VMD or
GaussView, but I wanted to take advantage of Mathematicas capability to easily combine graphics, as well as the ability to automate the process in order to efficiently create frames for a
movie.
First off, we need a function to extract the data from the cube file. In the process, we will create the text for an XYZ file, a format also developed by Gaussian. The function
OutForm is used here to mimic the printf function found in other programming languages.
OutForm[num_?NumericQ, width_Integer, ndig_Integer,
OptionsPattern[]] :=
Module[{mant, exp, val},
{mant, exp} = MantissaExponent[num];
mant = ToString[NumberForm[mant, {ndig, ndig}]];
exp = If[Sign[exp] == -1, "-", "+"] <> IntegerString[exp, 10, 2];
val = mant <> "E" <> exp;
StringJoin@PadLeft[Characters[val], width, " "]
];
ReadCube[cubeFileName_?StringQ] := Module[
{moltxt, nAtoms, lowerCorner, nx, ny, nz, xstep, ystep, zstep,
atoms, desc1, desc2, xyzText, cubeDat, xgrid, ygrid, zgrid,
dummy1, dummy2, atomicNumber, atomx, atomy, atomz, tmpString,
headerTxt,bohr2angstrom},
bohr2angstrom = 0.529177249;
moltxt = OpenRead[cubeFileName];
desc1 = Read[moltxt, String];
desc2 = Read[moltxt, String];
lowerCorner = {0, 0, 0};
{nAtoms, lowerCorner[[1]], lowerCorner[[2]], lowerCorner[[3]]} =
Read[moltxt, String] // ImportString[#, "Table"][[1]] &;
xyzText = ToString[nAtoms] <> "\n";
xyzText = xyzText <> desc1 <> desc2 <> "\n";
{nx, xstep, dummy1, dummy2} =
Read[moltxt, String] // ImportString[#, "Table"][[1]] &;
{ny, dummy1, ystep, dummy2} =
Read[moltxt, String] // ImportString[#, "Table"][[1]] &;
{nz, dummy1, dummy2, zstep} =
Read[moltxt, String] // ImportString[#, "Table"][[1]] &;
Do[
{atomicNumber, dummy1, atomx, atomy, atomz} =
Read[moltxt, String] // ImportString[#, "Table"][[1]] &;
xyzText = If[Sign[lowerCorner[[1]]] == 1,
xyzText <> ElementData[atomicNumber, "Abbreviation"] <>
OutForm[atomx, 17, 7] <> OutForm[atomy, 17, 7] <>
OutForm[atomz, 17, 7] <> "\n",
xyzText <> ElementData[atomicNumber, "Abbreviation"] <>
OutForm[bohr2angstrom atomx, 17, 7] <>
OutForm[bohr2angstrom atomy, 17, 7] <>
OutForm[bohr2angstrom atomz, 17, 7] <> "\n"];
, {nAtoms}];
cubeDat =
Partition[Partition[ReadList[moltxt, Number, nx ny nz], nz], ny];
Close[moltxt];
moltxt = OpenRead[cubeFileName];
headerTxt = Read[moltxt, Table[String, {2 + 4 + nAtoms}]];
Close[moltxt];
headerTxt = StringJoin@Riffle[headerTxt, "\n"];
xgrid =
Range[lowerCorner[[1]], lowerCorner[[1]] + xstep (nx - 1), xstep];
ygrid =
Range[lowerCorner[[2]], lowerCorner[[2]] + ystep (ny - 1), ystep];
zgrid =
Range[lowerCorner[[3]], lowerCorner[[3]] + zstep (nz - 1), zstep];
{cubeDat, xgrid, ygrid, zgrid, xyzText, headerTxt}
];
If you need to create a cube file, then the following function can be used:
WriteCube[cubeFileName_?StringQ, headerTxt_?StringQ, cubeData_] :=
Module[{stream},
stream = OpenWrite[cubeFileName, FormatType -> FortranForm];
WriteString[stream, headerTxt, "\n"];
Map[WriteString[stream, ##, "\n"] & @@
Riffle[ScientificForm[#, {3, 4},
NumberFormat -> (Row[{#1, "E", If[#3 == "", "+00", #3],
"\t"}] &), NumberPadding -> {"", "0"},
NumberSigns -> {"-", " "}] & /@ #, "\n", {7, -1, 7}] &,
cubeData, {2}];
Close[stream];]
Next we need the function to plot the orbital,
CubePlot[{cub_, xg_, yg_, zg_, xyz_}, plotopts : OptionsPattern[]] :=
Module[{xyzplot, bohr2picometer, datarange3D, pr},
bohr2picometer = 52.9177249;
datarange3D =
bohr2picometer {{xg[[1]], xg[[-1]]}, {yg[[1]],
yg[[-1]]}, {zg[[1]], zg[[-1]]}};
xyzplot = ImportString[xyz, "XYZ"];
Show[xyzplot,
ListContourPlot3D[Transpose[cub, {3, 2, 1}],
Evaluate[FilterRules[{plotopts}, Options[ListContourPlot3D]]],
Contours -> {-.02, .02}, ContourStyle -> {Blue, Red},
DataRange -> datarange3D, MeshStyle -> Gray,
Lighting -> {{"Ambient", White}}],
Evaluate[
FilterRules[{plotopts}, {ViewPoint, ViewVertical, ImageSize}]]]
];
Lets look at an example. First we need to read in a cube file, download this cube file and place it in your base directory:
cys-MO35cube{cubedata,xg,yg,zg,xyz,header}= ReadCube["cys-MO35.cube"];
Then plot it via
CubePlot[{cubedata, xg, yg, zg, xyz}]
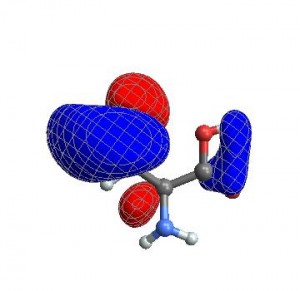
When I want to create a movie file, I want all the images to have exactly the same
ViewAngle,
ViewPoint, and
ViewCenter. When you give these options to
CubePlot, it feeds them directly to the
Show function
vp = {ViewCenter -> {0.5, 0.5, 0.5},
ViewPoint -> {1.072, 0.665, -3.13},
ViewVertical -> {0.443, 0.2477, 1.527}};
CubePlot[{cubedata, xg, yg, zg, xyz}, vp]
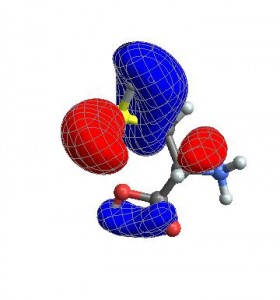
Finally, you can also give any options that normally go to
ListContourPlot3DCubePlot[{cubedata, xg, yg, zg, xyz}, vp,
ContourStyle -> {Texture[ExampleData[{"ColorTexture", "Vavona"}]],
Texture[ExampleData[{"ColorTexture", "Amboyna"}]]},
Contours -> {-.015, .015}]

Many thanks to Daniel Healion for the
ReadCube and
WriteCube functions.