Hi Sotirios,
here comes a simple first guess:
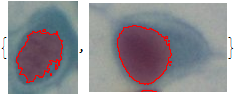
ClearAll["Global`*"]
web = {"http://community.wolfram.com//c/portal/getImageAttachment?\
filename=cell-4-9-1.jpg&userId=1438211",
"http://community.wolfram.com//c/portal/getImageAttachment?\
filename=cell-4-9-2.jpg&userId=1438211"};
imgs0 = Import /@ web;
thrh = FindThreshold /@ imgs0;
nucs = ColorNegate /@ MapThread[MorphologicalBinarize, {imgs0, thrh}];
MapThread[HighlightImage, {imgs0, nucs}]
This gives:
Does that help?
Regards -- Henrik