Dear Veit,
I know nothing about pdb, but perhaps this hack may be a useful direction for you? Here a GraphicsComplex is deconstructed to extract information.
example =
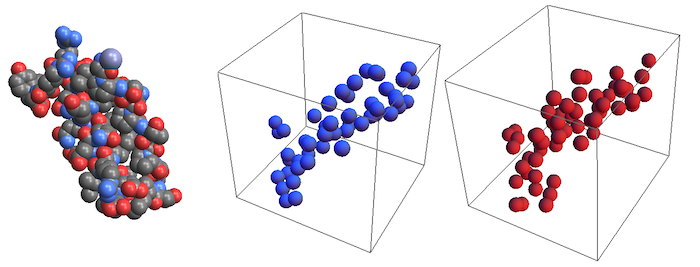
Import["ExampleData/1PPT.pdb", "Rendering" -> "Spacefilling"]
centers = Cases[example, GraphicsComplex[a_, __] :> a, Infinity][[1]]
spheres = Cases[example, GraphicsComplex[a_, b_] :> b, Infinity][[1]]
types = Partition[SplitBy[spheres, (Head[#] == RGBColor) &], 2]
typeColors = Flatten[First /@ types]
typeCoordinates =
Cases[#, Sphere[coord_, _] :> centers[[coord]], Infinity] & /@ types
(*examples*)
typeColors[[2]]
typeCoordinates[[2]]
Graphics3D[{typeColors[[2]],
Sphere[#, 150] & /@ typeCoordinates[[2]]}]
Graphics3D[{typeColors[[3]],
Sphere[#, 150] & /@ typeCoordinates[[3]]}]