Hi Antonio, How this happens depends on which plotting function you use to display the image. Mathematica ListPlot and ContourPlot and similar functions plot f[x,y] with x on the horizontal and y on the vertical, increasing left to right and bottom to top as usual. But ArrayPlot and similar functions plot array[[row,column]] with rows on the vertical axis, increasing top to bottom, and columns on the horizontal, increasing left to right. (Edit: It looks like the graphic of an image, as when it is first imported, is oriented like the graphic plotting routines, not like ArrayPlot.)
So if we build an interpolating function with the row and column indices in the x and y position we get this rotation, and ListInterpolation will do this.
temp = {
{1, 1, 0, 0, 0},
{1, 1, 0, 0, 0},
{0, 0, 0, 0, 0},
{0, 0, 0, 0, 0},
{0, 0, 0, 0, 0}};
tf = ListInterpolation[temp];
ContourPlot[tf[x, y], {x, 1, 5}, {y, 1, 5}, PlotLegends -> Automatic]
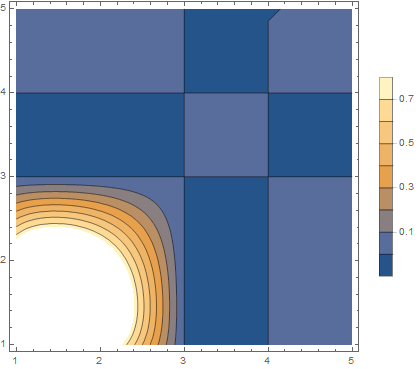
This could be overcome by building the Interpolating function differently. Also, ArrayPlot has an option that helps. (Note that ArrayPlot inverts intensity to create a negative image.) Here Transpose swaps rows for columns, and DataReversed plots bottom to top.
ArrayPlot[temp // Transpose, DataReversed -> True]

Best,
David