Introduction
This post has links and code that demonstrate the use of classifier agnostic procedure for finding the importance of variables in labeled data. (The values of which variables are most decisive in the implication of labels.)
The procedure was used in this blog Classification and association rules for census income data. It is implemented in the package "VariableImportanceByClassifiers.m". I took it from [5] and it is (briefly) described also below.
The document Importance of variables investigation guide, [3], has much more extensive descriptions, explanations, and code for importance of variables investigation using classifiers, Mosaic plots, Decision trees, Association rules, and Dimension reduction
This post parallels and completes the WordPress.com blog post "Importance of variables investigation".
Procedure outline
Here we describe the procedure used (see [5]).
Split the data into training and testing datasets.
Build a classifier with the training set.
Verify using the test set that good classification results are obtained. Find the baseline accuracy.
If the number of variables (attributes) is $k$ for each $i$, $1\leq i\leq k$:
4.1 Shuffle the values of the $i$-th column of the test data and find the classification success rates.
Compare the obtained $k$ classification success rates between each other and with the success rates obtained by the un-shuffled test data.
The variables for which the classification success rates are the worst are the most decisive.
Note that instead of using the overall baseline accuracy we can make the comparison over the accuracies for selected, more important class labels. (See the examples below.)
The procedure is classifier agnostic. With certain classifiers, Naive Bayesian classifiers and Decision trees, the importance of variables can be directly concluded from their structure obtained after training.
The procedure can be enhanced by using dimension reduction before building the classifiers. (See [3] for an outline.)
Implementation description
The implementation of the procedure is straightforward in Mathematica -- see the package VariableImportanceByClassifiers.m.
The package can be imported with the command:
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/VariableImportanceByClassifiers.m"]
At this point the package has only one function, AccuracyByVariableShuffling, that takes as arguments a ClassifierFunction object, a dataset, optional variable names, and the option "FScoreLabels" that allows the use of accuracies over a custom list of class labels instead of overall baseline accuracy.
Here is the function signature:
AccuracyByVariableShuffling[ clFunc_ClassifierFunction, testData_, variableNames_:Automatic, opts:OptionsPattern[] ]
The returned result is an Association structure that contains the baseline accuracy and the accuracies corresponding to the shuffled versions of the dataset. I.e. steps 3 and 4 of the procedure are performed by AccuracyByVariableShuffling. Returning the result in the form Association[___] means we can treat the result as a list with named elements similar to the list structures in Lua and R.
For the examples in the next section we also going to use the package MosaicPlot.m, that can be imported with the following command:
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/MosaicPlot.m"]
Concrete application over the "Mushroom" dataset
.1. Load some data.
testSetName = "Mushroom"; (* "Titanic" *)
trainingSet = ExampleData[{"MachineLearning", testSetName}, "TrainingData"];
testSet = ExampleData[{"MachineLearning", testSetName}, "TestData"];
.2. Variable names and unique class labels.
varNames = Flatten[List @@ ExampleData[{"MachineLearning", testSetName}, "VariableDescriptions"]]
(* {"cap-shape", "cap-surface", "cap-color", "bruises?", "odor", \
"gill-attachment", "gill-spacing", "gill-size", "gill-color", "stalk-shape", \
"stalk-root", "stalk-surface-above-ring", "stalk-surface-below-ring", \
"stalk-color-above-ring", "stalk-color-below-ring", "veil-type", \
"veil-color", "ring-number", "ring-type", "spore-print-color", "population", \
"habitat", "edibility of mushroom (either edible or poisonous)"} *)
The class labels are:
classLabels = Union[ExampleData[{"MachineLearning", testSetName}, "Data"][[All, -1]]]
(* {"edible", "poisonous"} *)
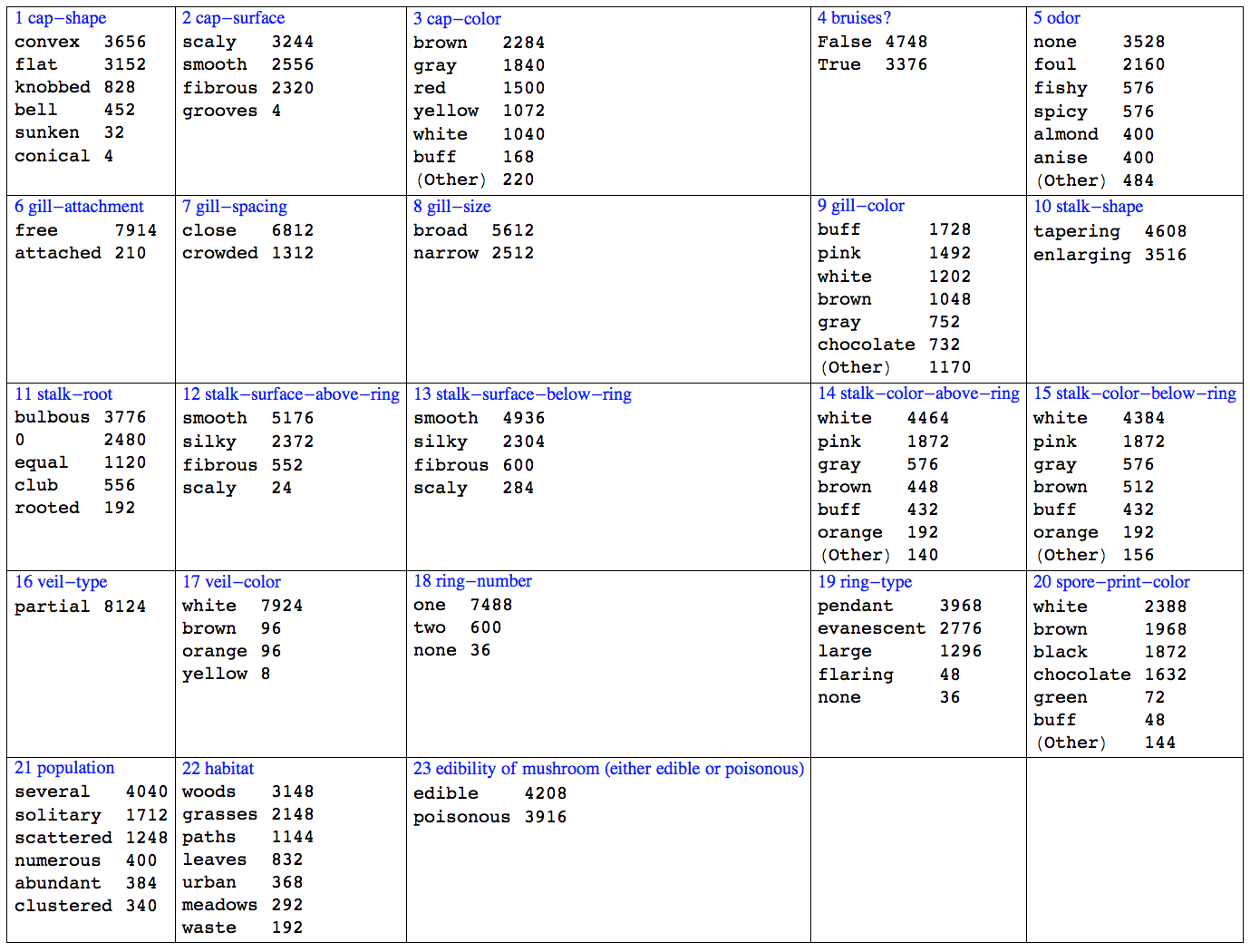
.3. Here is a data summary.
Grid[ArrayReshape[
RecordsSummary[(Flatten /@ (List @@@
Join[trainingSet, testSet])) /. _Missing -> 0, varNames], {5,
5}, ""], Dividers -> All, Alignment -> {Left, Top}]
.4. Make the classifier.
clFunc = Classify[trainingSet, Method -> "RandomForest"]
.5. Obtain accuracies after shuffling.
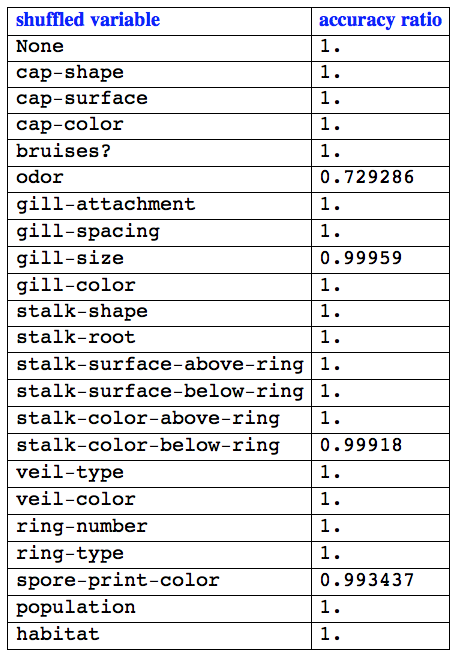
accs = AccuracyByVariableShuffling[clFunc, testSet, varNames]
(* <|None -> 1., "cap-shape" -> 1., "cap-surface" -> 1., "cap-color" -> 1.,
"bruises?" -> 1., "odor" -> 0.729286, "gill-attachment" -> 1.,
"gill-spacing" -> 1., "gill-size" -> 0.99959, "gill-color" -> 1.,
"stalk-shape" -> 1., "stalk-root" -> 1., "stalk-surface-above-ring" -> 1.,
"stalk-surface-below-ring" -> 1., "stalk-color-above-ring" -> 1.,
"stalk-color-below-ring" -> 0.99918, "veil-type" -> 1., "veil-color" -> 1.,
"ring-number" -> 1., "ring-type" -> 1., "spore-print-color" -> 0.993437,
"population" -> 1., "habitat" -> 1.|> *)
.6. Tabulate the results.
Grid[
Prepend[
List @@@ Normal[accs/First[accs]],
Style[#, Bold, Blue,
FontFamily -> "Times"] & /@ {"shuffled variable", "accuracy ratio"}],
Alignment -> Left, Dividers -> All]
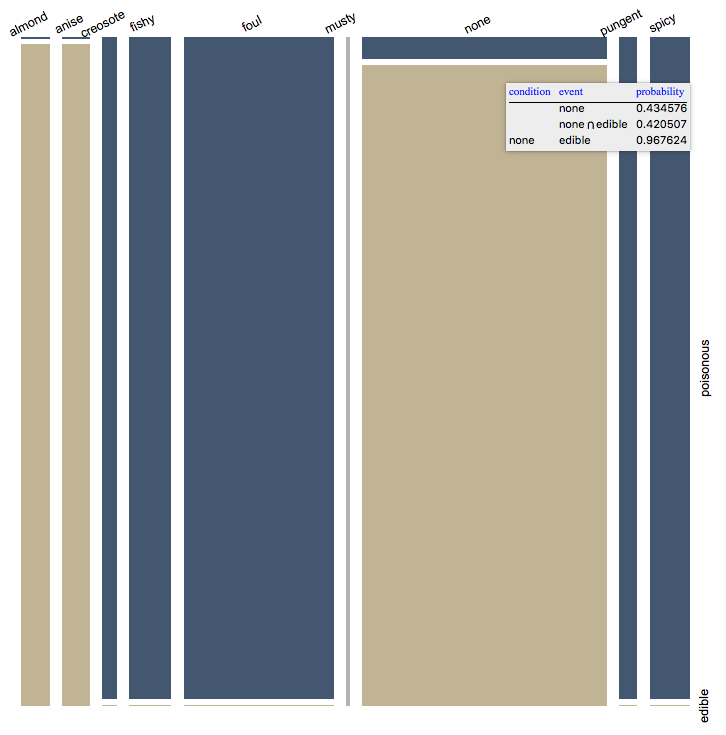
.7. Further confirmation of the found variable importance can be done using the mosaic plots. Looking at the plot we can see why "odor" is so decisive -- the odor values for "poisonous" and "edible" intersect very little.
t = (Flatten /@ (List @@@ trainingSet));
MosaicPlot[t[[All, {5, -1}]], "FirstAxis" -> "Top",
"LabelRotation" -> {{1, 0.5}, {0, 1}},
ColorRules -> {2 -> ColorData[7, "ColorList"]} ]
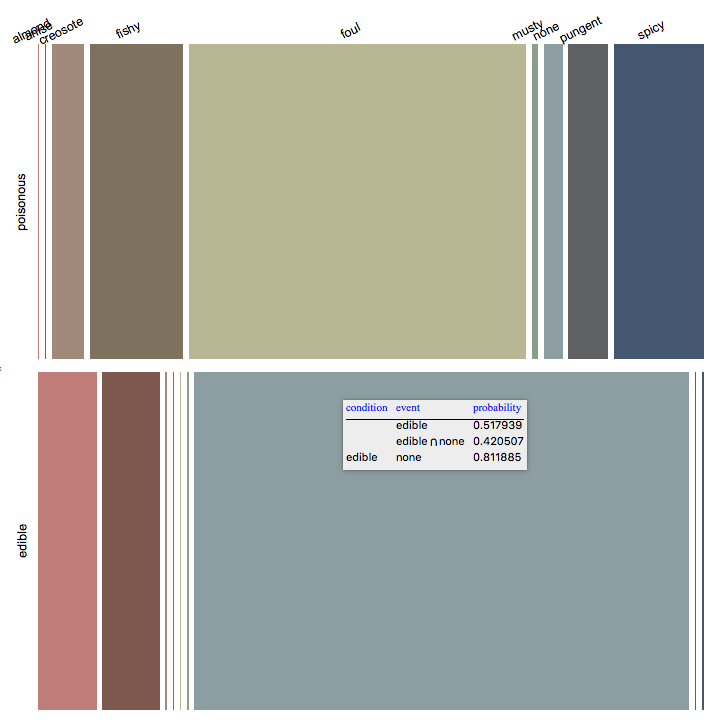
Here is a mosaic plot showing the conditional probabilities from a different direction.
.5a. In order to use F-scores instead of overall accuracy the desired class labels are specified with the option "FScoreLabels".
accs = AccuracyByVariableShuffling[clFunc, testSet, varNames, "FScoreLabels" -> classLabels]
(* <|None -> {1., 1.}, "cap-shape" -> {1., 1.}, "cap-surface" -> {1., 1.},
"cap-color" -> {1., 1.}, "bruises?" -> {1., 1.},
"odor" -> {0.677028, 0.763933}, "gill-attachment" -> {1., 1.},
"gill-spacing" -> {1., 1.}, "gill-size" -> {0.999209, 0.999148},
"gill-color" -> {1., 1.}, "stalk-shape" -> {1., 1.},
"stalk-root" -> {1., 1.}, "stalk-surface-above-ring" -> {0.999209, 0.999148},
"stalk-surface-below-ring" -> {1., 1.},
"stalk-color-above-ring" -> {1., 1.}, "stalk-color-below-ring" -> {1., 1.},
"veil-type" -> {1., 1.}, "veil-color" -> {1., 1.}, "ring-number" -> {1., 1.},
"ring-type" -> {1., 1.}, "spore-print-color" -> {0.995249, 0.994894},
"population" -> {1., 1.}, "habitat" -> {1., 1.}|> *)
.5b. Here is another example that uses the class label with the smallest F-score. (Probably the most important since it is most mis-classified).
accs = AccuracyByVariableShuffling[clFunc, testSet, varNames,
"FScoreLabels" -> Position[#, Min[#]][[1, 1, 1]] &@
ClassifierMeasurements[clFunc, testSet, "FScore"]]
(* <|None -> {1.}, "cap-shape" -> {1.}, "cap-surface" -> {1.},
"cap-color" -> {1.}, "bruises?" -> {1.}, "odor" -> {0.679043},
"gill-attachment" -> {1.}, "gill-spacing" -> {1.}, "gill-size" -> {0.999604},
"gill-color" -> {1.}, "stalk-shape" -> {1.}, "stalk-root" -> {1.},
"stalk-surface-above-ring" -> {0.999209}, "stalk-surface-below-ring" -> {1.},
"stalk-color-above-ring" -> {1.}, "stalk-color-below-ring" -> {1.},
"veil-type" -> {1.}, "veil-color" -> {1.}, "ring-number" -> {1.},
"ring-type" -> {1.}, "spore-print-color" -> {0.994851}, "population" -> {1.},
"habitat" -> {1.}|> *)
.5c. It is good idea to verify that we get the same results using different classifiers. Below is given code that computes the shuffled accuracies and returns the relative damage scores for a set of methods of Classify.
mres = Association@Map[
Function[{clMethod},
cf = Classify[trainingSet, Method -> clMethod];
accRes = AccuracyByVariableShuffling[cf, testSet, varNames];
clMethod -> (accRes[None] - Rest[accRes])/accRes[None]
], {"LogisticRegression", "NearestNeighbors", "NeuralNetwork",
"RandomForest", "SupportVectorMachine"}] ;
Magnify[#, 0.8] &@Dataset[mres]

References
1] Anton Antonov, [Classification and association rules for census income data, (2014), MathematicaForPrediction at WordPress.com.
2] Anton Antonov, [Variable importance determination by classifiers implementation in Mathematica, (2015), source code at MathematicaForPrediction at GitHub, package VariableImportanceByClassifiers.m.
3] Anton Antonov, [Importance of variables investigation guide, (2016), MathematicaForPrediction at GitHub, folder Documentation.
4] Anton Antonov, [Mosaic plot for data visualization implementation in Mathematica, (2014), MathematicaForPrediction at GitHub, package MosaicPlot.m.
5] Leo Breiman et al., [Classification and regression trees, Chapman & Hall, 1984, ISBN-13: 978-0412048418.