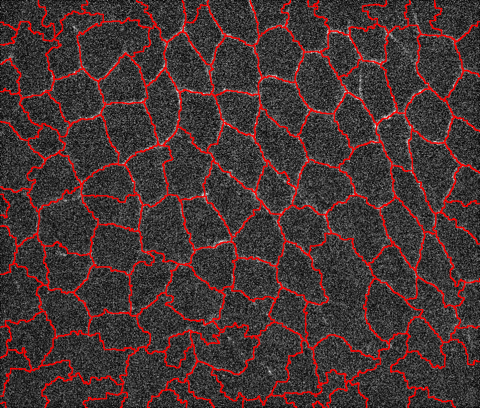
Recently we had a question about a noisy microscope image of animal tissue, see image below. The goal was to get the contours for all biological cells boundaries. At the bottom image fades significantly but the human eye can still detect some contours. One trick that generally works is to attempt to find the valleys instead of the lines and then segment the image to find the lines separating the valleys. Let's import image first:
i = Import["https://wolfr.am/gqaPy6GP"]
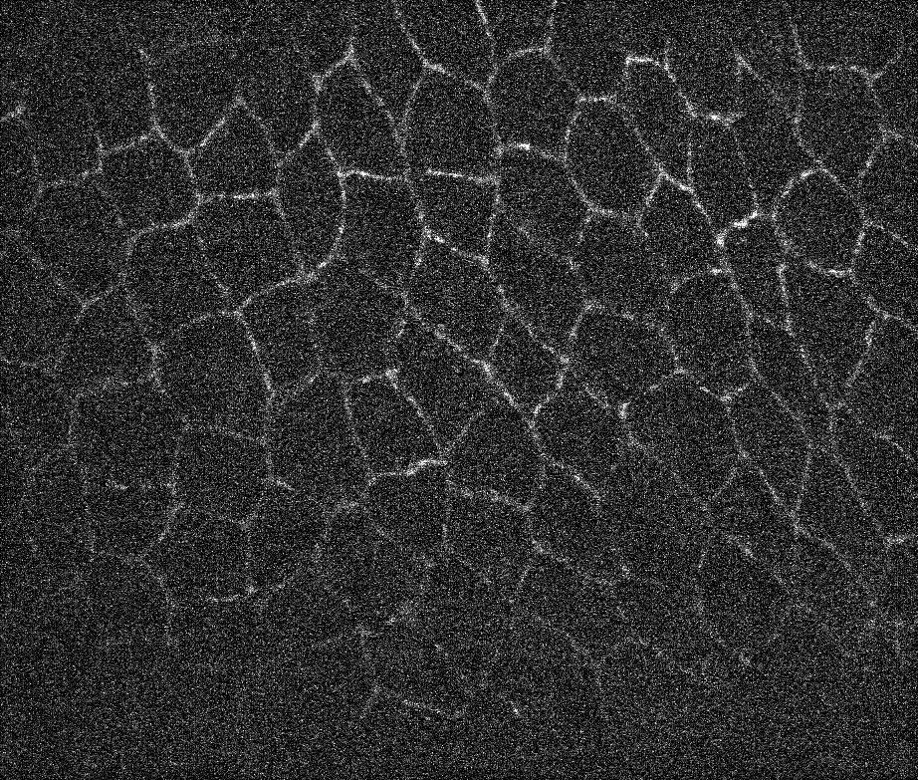
Here is one way you can fairly robustly find the centers of the cells:
marker = MaxDetect[ImageAdjust@LaplacianGaussianFilter[i, 50], .05];
HighlightImage[i, marker]
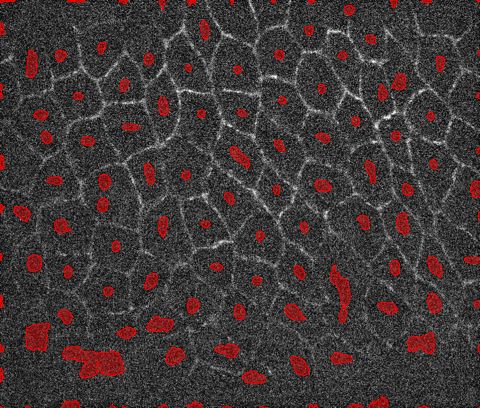
Once you have a good estimate of the cell centers, you can run a segmentation algorithm like watershed using the markers. Here I am segmenting a slightly smoothed version of the image using MedianFilter:
ws = WatershedComponents[MedianFilter[i, 4], marker];
Colorize[ws]
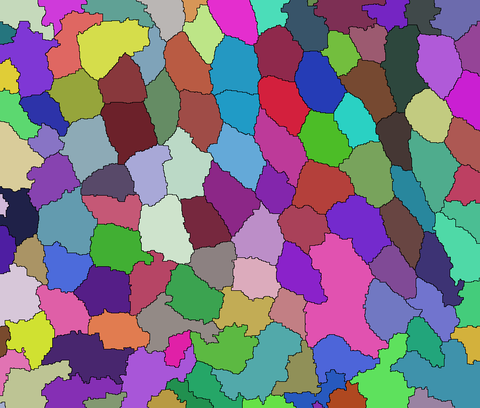
And, then extract the edges separating detected components:
boundaries = ColorNegate[Image[ws, "Bit"]];
HighlightImage[i, boundaries]