Wolfram Language has very detailed built-in data about Anatomical Structure. Here is an example of usage building an interactive models of brain. First we would need some data. Because anatomical parts constitute of smaller parts and so forth, we can visualize anatomical structures as trees, for instance:
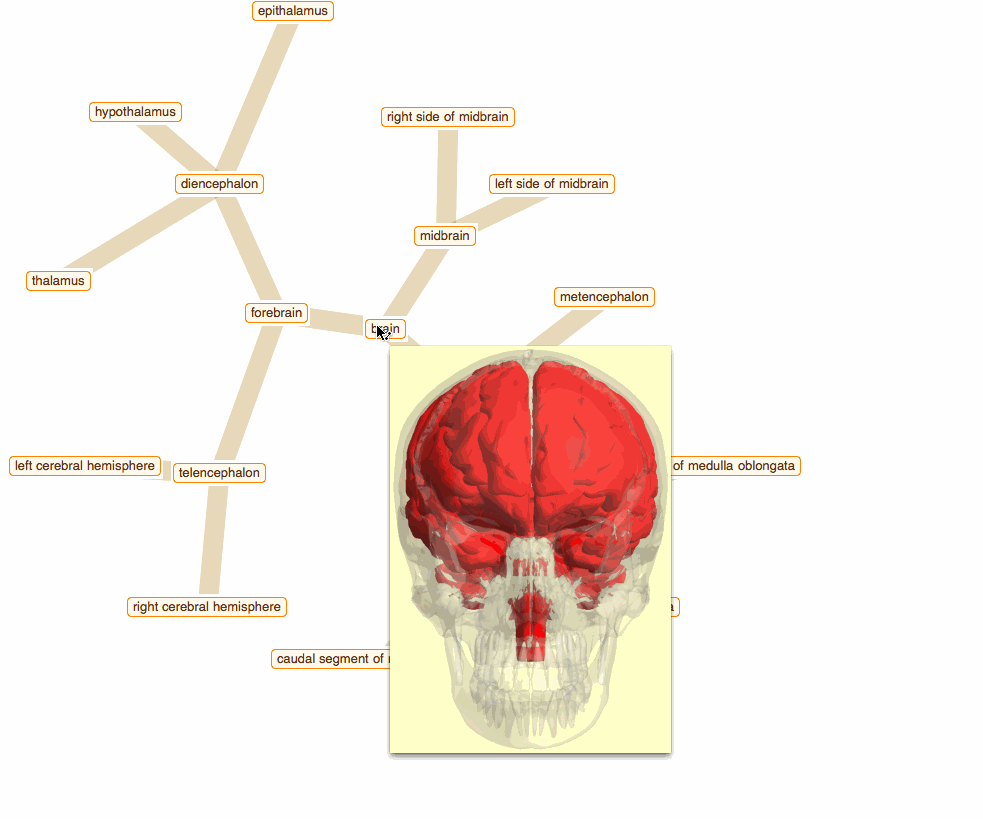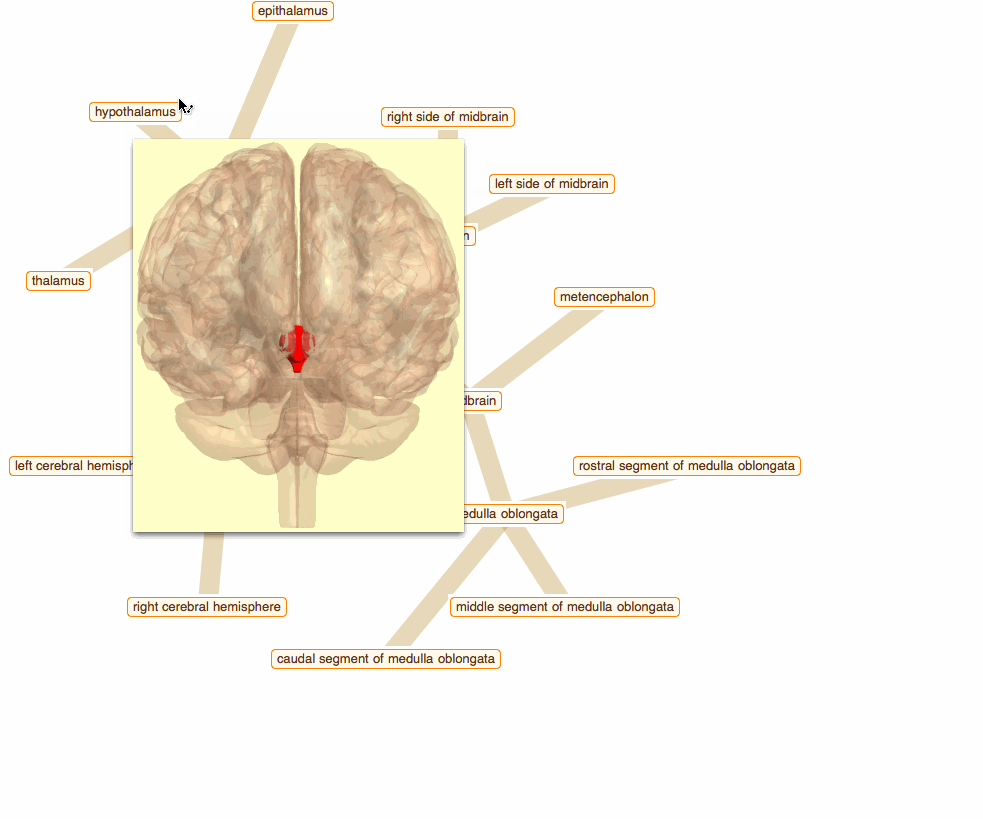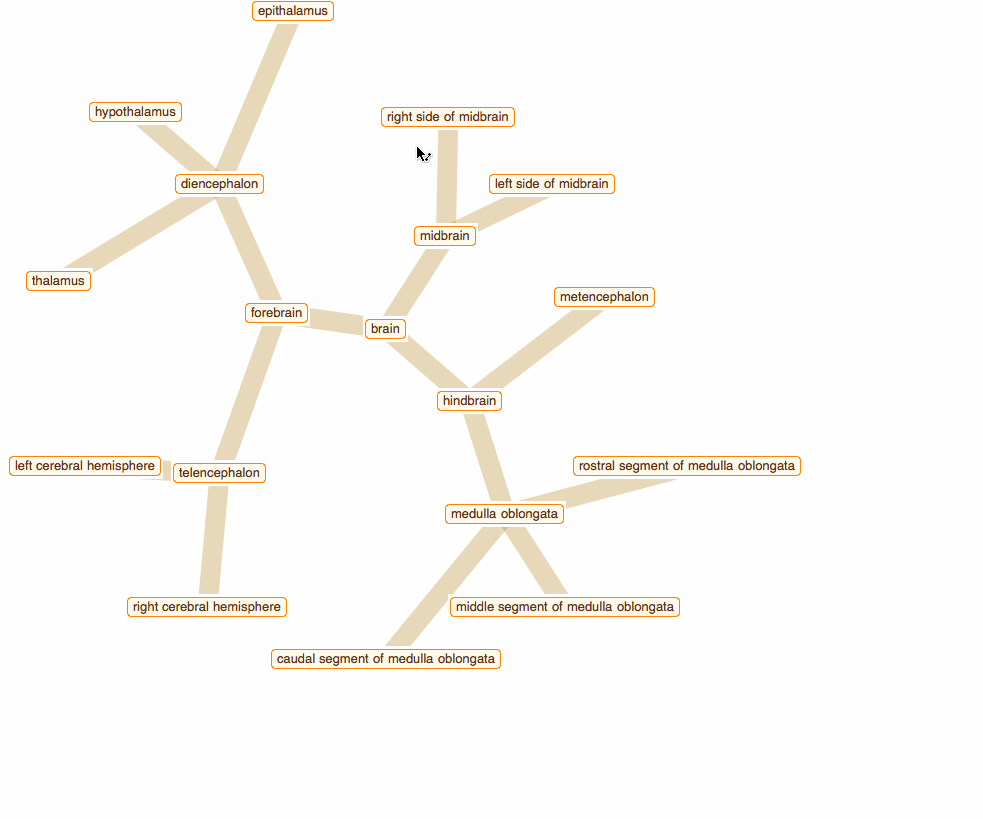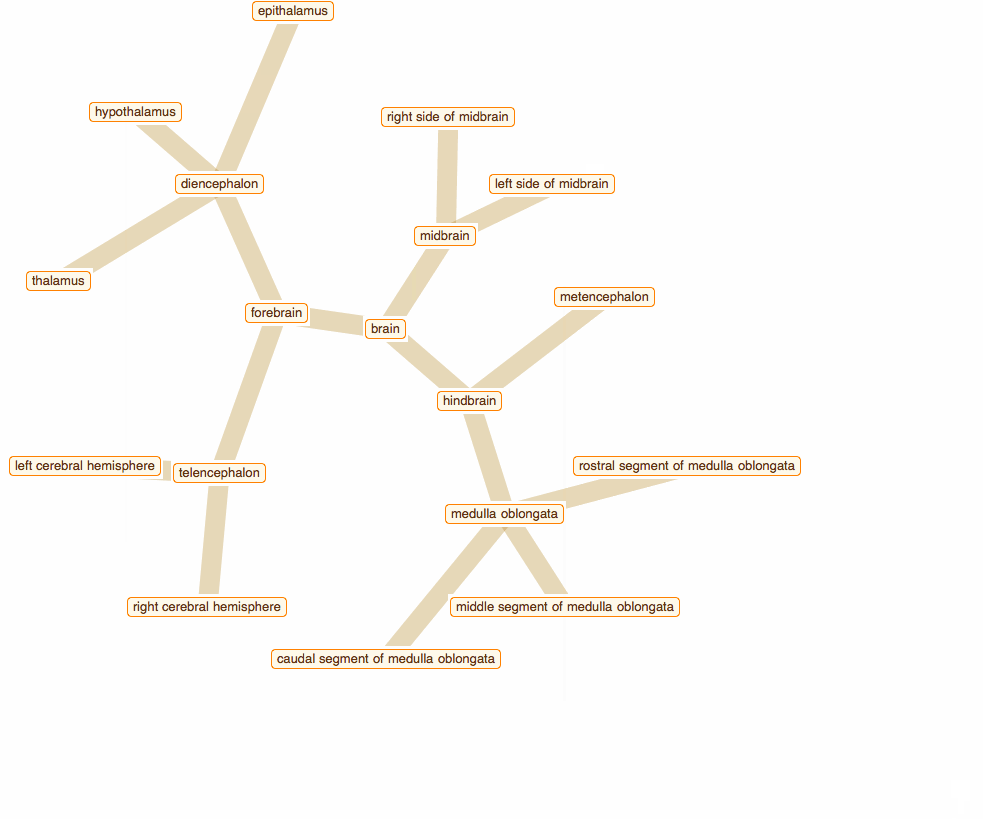
To get that I need to build a data crawler:
crawler[root_, depth_] :=
Flatten[Rest[NestList[DeleteCases[Union[Flatten[
Thread[# -> DeleteMissing[EntityValue[#, EntityProperty["AnatomicalStructure",
"RegionalParts"]]]] & /@ Last /@ #]], HoldPattern[_ -> Sequence[]]] &, {"" -> root}, depth]]];
start from brain and get down 3 levels of anatomical structure:
edge = crawler[Entity["AnatomicalStructure", "Brain"], 3]
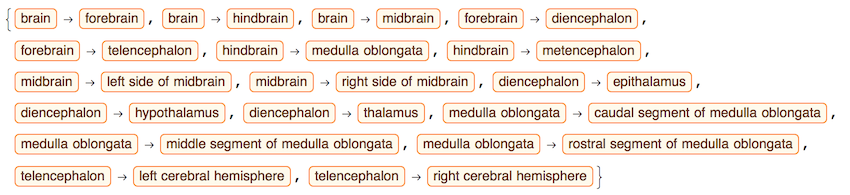
Get edges, vertices, and labels as images of anatomical parts:
g = Graph[edge];
vert = VertexList[g];
lblsT = Rasterize /@ vert;
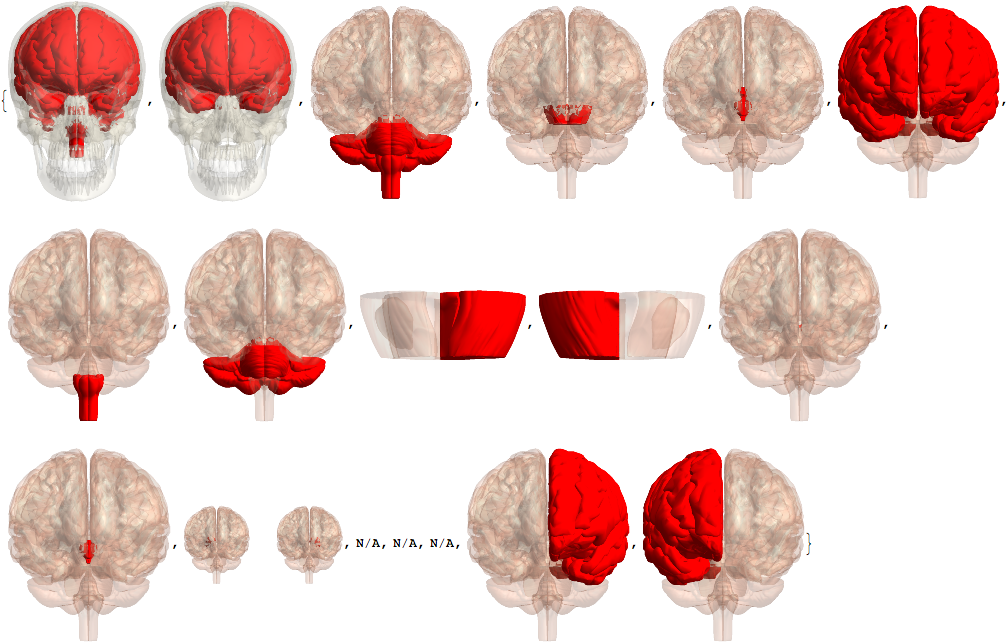
The image at the top is build with ContextualizedHighlightedImage:
lblsI = Module[{tmp},
tmp = EntityValue[#, EntityProperty["AnatomicalStructure", "ContextualizedHighlightedImage"]];
If[ImageQ[tmp], ImageCrop[tmp], "N/A"]] & /@ vert
we can also try just with Image:
lblsI = Module[{tmp},
tmp = EntityValue[#, EntityProperty["AnatomicalStructure", "Image"]];
If[ImageQ[tmp], ImageCrop[tmp], "N/A"]] & /@ vert
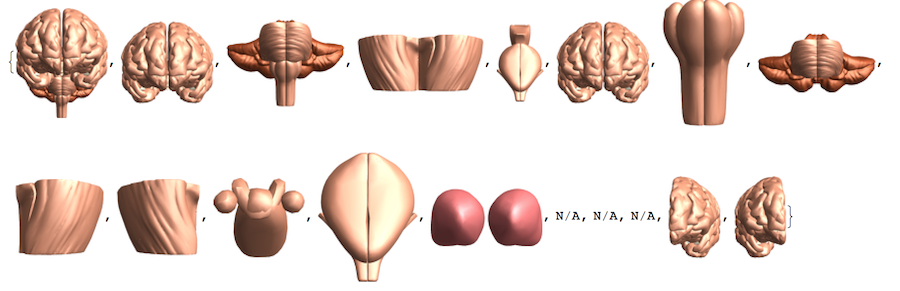
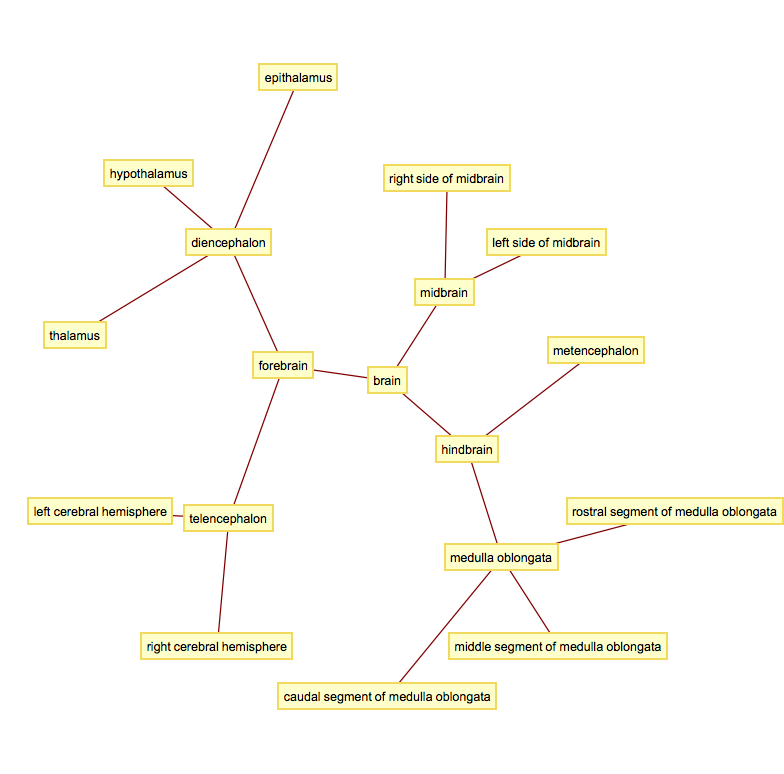
A quick way to get a non-interactive tree like plot:
GraphPlot[edge, VertexLabeling -> True]
But for fancy interactivity and to preserve Graph data-structure we write a short code and get the top nice image in this post:
tltps = Tooltip @@@ Transpose[{lblsT, lblsI}];
TreeGraph[vert, edge, VertexLabels -> Thread[vert -> (Placed[#, Center] & /@ tltps)],
ImageSize -> 700, GraphStyle -> "ThickEdge", GraphLayout -> "RadialEmbedding"]
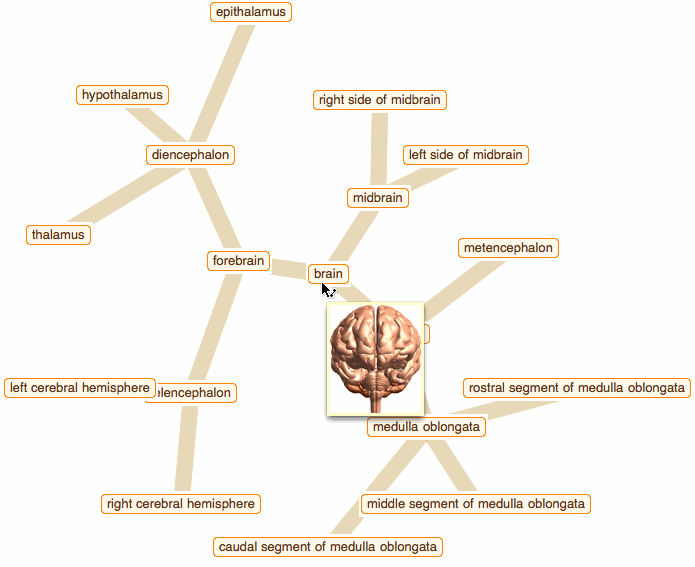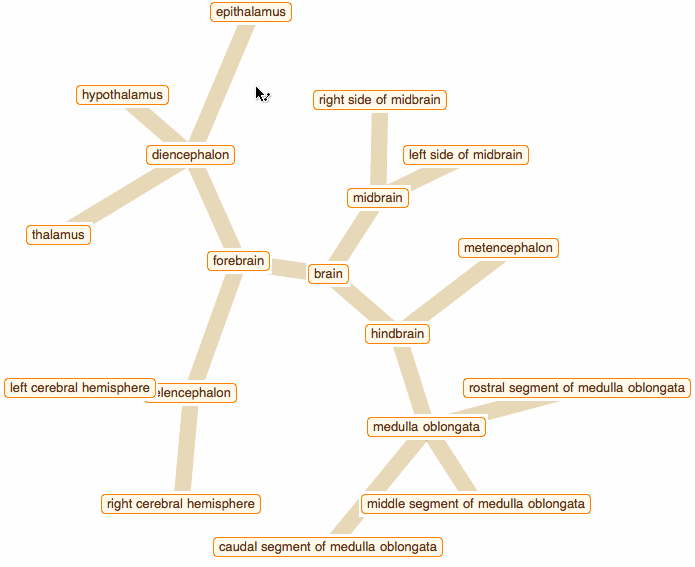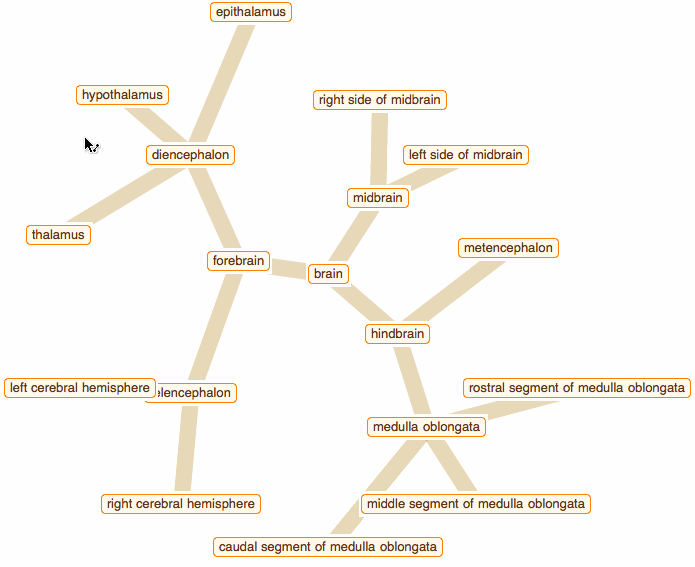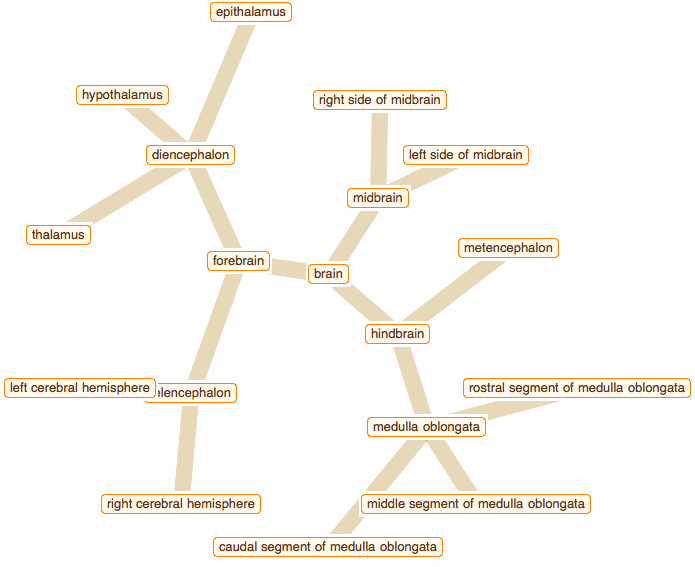
That was a good programming exercise and now we can appreciate how recent function NestGraph can make things much easier:
NestGraph[Cases[EntityValue[#, EntityProperty["AnatomicalStructure", "RegionalParts"]], _Entity] &,
Entity["AnatomicalStructure", "Brain"], 4, ImageSize -> 700, GraphStyle -> "SmallNetwork",
GraphLayout -> "RadialEmbedding", VertexLabels -> Placed["Name", Tooltip]]
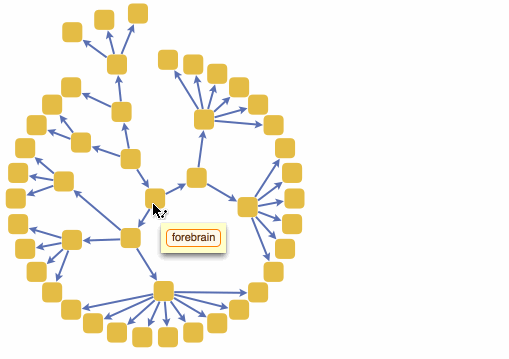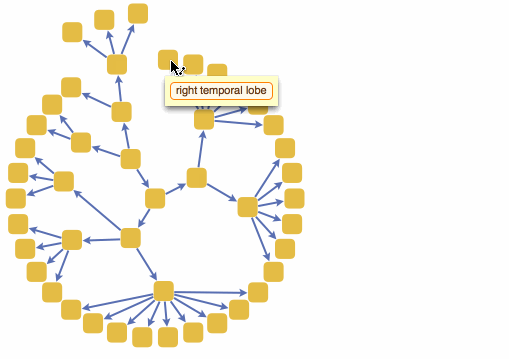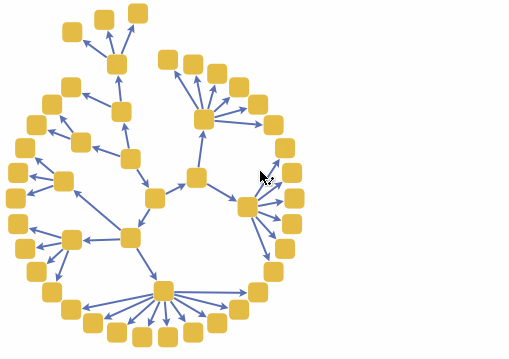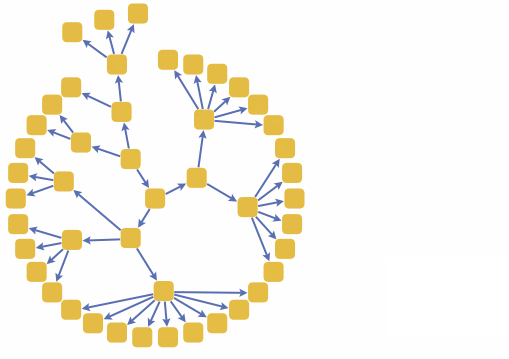
We can also build a 3D interactive model app. For example, choose specific brain parts
bParts = {
Entity["AnatomicalStructure", "ParietalLobe"],
Entity["AnatomicalStructure", "Cerebellum"],
Entity["AnatomicalStructure", "PituitaryGland"],
Entity["AnatomicalStructure", "OpticChiasm"],
Entity["AnatomicalStructure", "Hypothalamus"],
Entity["AnatomicalStructure", "ChoroidPlexusOfCerebralHemisphere"],
Entity["AnatomicalStructure", "HippocampalFormation"]}

and get their 3D models:
bParts3D = EntityValue[bParts, EntityProperty["AnatomicalStructure", "Graphics3D"]];
And then we can build an app with just few lines of code:
Manipulate[
Show[Graphics3D[], x, ImageSize -> {400, 400},
SphericalRegion -> True, Boxed -> False, ViewAngle -> .27],
{{x, bParts3D[[1]], ""}, Thread[bParts3D -> bParts],
CheckboxBar, Appearance -> "Vertical"}]
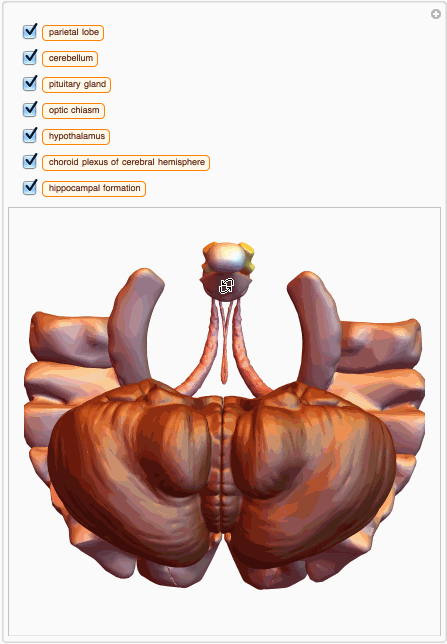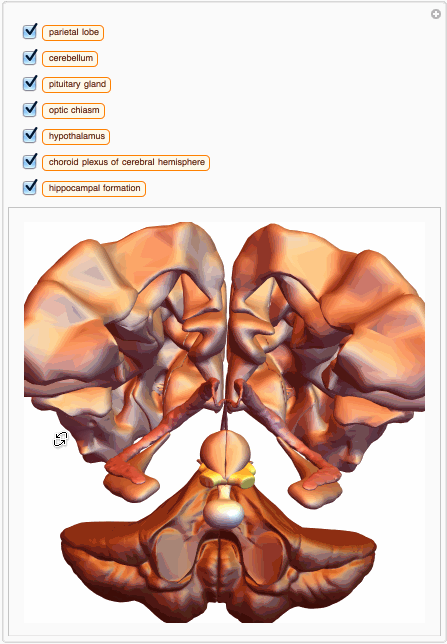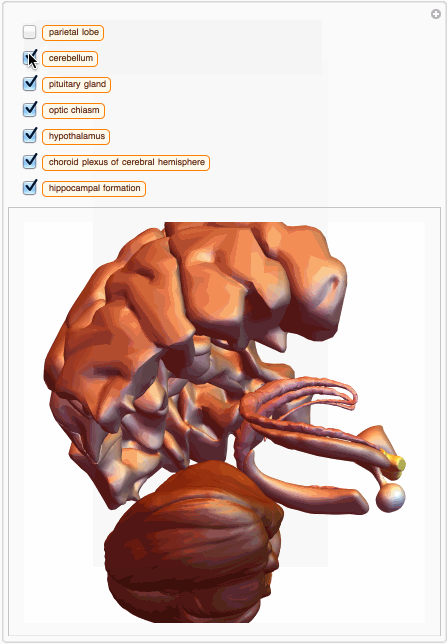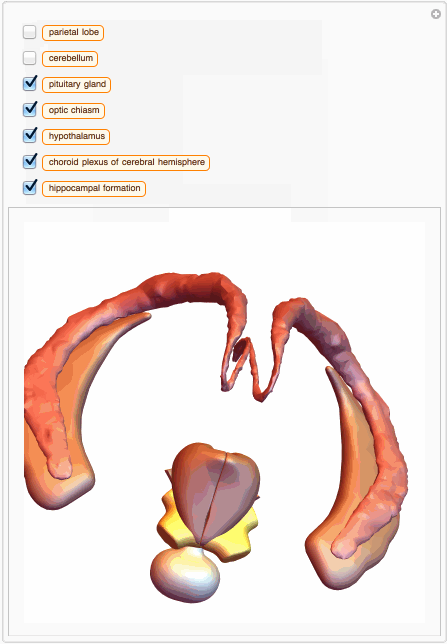
You can also get MeshRegion
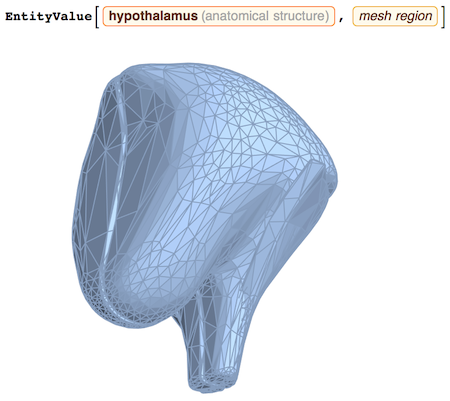
to do some computational geometry, for example:
RegionMeasure[%]
1482.164538484
Of course you can check for any anatomical part and also its properties:
EntityProperties["AnatomicalStructure"]
